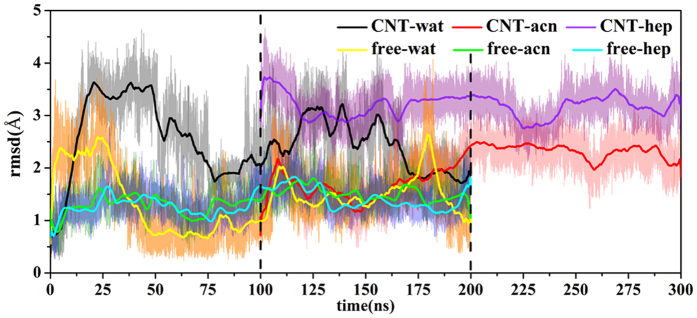
Figure 6. Changes in RMSD values of backbone atoms of two β-strands (Gly100-Tyr103, and Ser125-Gly128) with respect to the 200 ns simulation times for the free or the immobilized enzymes in the three different solvents.
The RMSD is deviation from the crystal structure. The initial structure of the enzyme-CNT complex in the two organic systems is derived from the final snapshot of the first 100 ns trajectory in CNT-wat system.