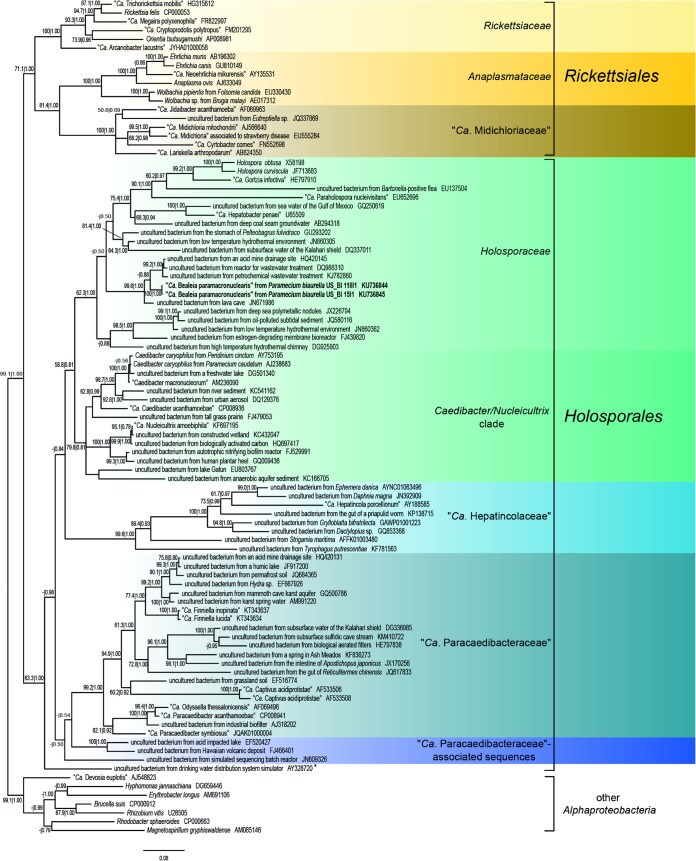
FIG 2.
Bayesian inference phylogenetic tree of “Candidatus Bealeia paramacronuclearis,” based on prokaryotic SSU rRNA gene sequences, including 18 Rickettsiales, 70 Holosporales, and 7 members of other orders of Alphaproteobacteria as an outgroup. The GTR+I+G model was employed. A total of 1,251 nucleotide columns were used to calculate the tree. Numbers indicate maximum likelihood bootstrap values after 1,000 pseudoreplicates and Bayesian posterior probabilities after 1,500,000 iterations. Values below 50% and 0.5 are not shown. Scale bar, 8 nucleotide substitutions per 100 positions. Ca., Candidatus. *, a sequence which clusters within “Candidatus Paracaedibacteraceae,” including associated sequences in the ML tree (see Fig. 3).