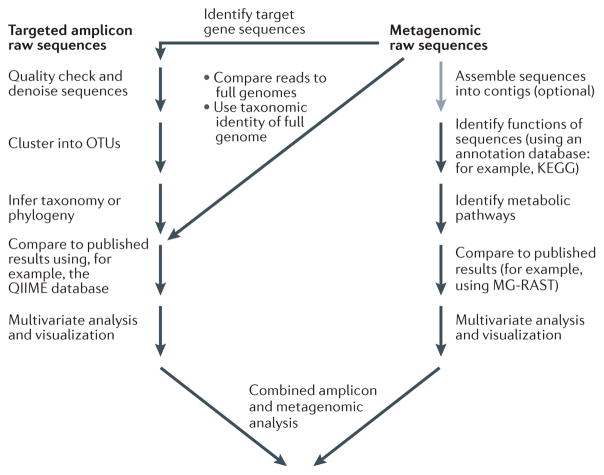
Figure 1. Bioinformatics analysis of microbiome sequence data.
Although variations exist, we show typical analysis paths for both targeted amplicon analysis (for example, analysis of 16S rDNA) and metagenomic analysis (for example, shotgun amplification). In targeted amplicon studies (left branch), raw sequences are usually passed through quality filtering and denoising algorithms to minimize the effects of sequencing artefacts. The resulting filtered sequence reads are clustered into operational taxonomic units (OTUs), representing similar organisms, and the phylogeny and taxonomic identity (when the organisms closely resemble named taxonomic groups) are inferred. At this stage, it is possible to incorporate sequence data from other relevant studies, or the data can be treated individually. The abundance of the various OTUs is then subjected to a variety of multivariate analyses and visualization procedures to elucidate the structure and patterns of the microbial communities. In metagenomic studies (right branch), the raw sequence fragments are sometimes assembled into contiguous sequences (contigs). The functional potential of those sequences is then typically assessed using a functional annotation database. The results are used to identify important metabolic pathways and are compared to the results of other metagenomic studies. The processed data are then subjected to multivariate analyses and visualizations, and they are often combined with the results of microbial profiling. Note that there are several opportunities for obtaining targeted gene data (similar to those produced in 16S rDNA gene surveys) in metagenomic studies, as indicated by the step labelled ‘identify target gene sequences’. KEGG, Kyoto Encyclopedia of Genes and Genomes; MG-RAST, Metagenomics Rapid Annotation using Subsystems Technology.