Figure 3.
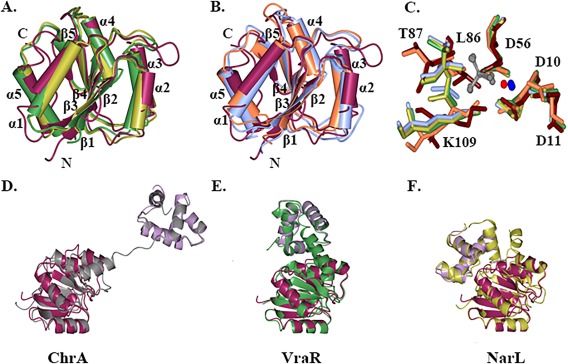
Structural comparison of RcsB with FixJ/NarL family regulators. A. Superposition of the nonphosphorylated RcsB receiver domain (maroon) with homologous receiver domain of YycF in inactive form (green) and PmrA in active form (gold). B. Superposition of the nonphosphorylated RcsB receiver domain with homologous receiver domain of CheY in inactive form (coral) and RR468 in active form (light blue). C. Superposition of conserved residues (D10, D11, D56, L86, T87, K109) at phosphorylation site of the RcsB receiver domain (tan) and active site residues of the receiver domain of RR468 with aspartate beryllium trifluoride residue (green) and magnesium ion, PmrA in complex with beryllium fluoride ion (carbons in light blue) and magnesium ion, CheY (orange) in complex with magnesium ion (red) and PhoP from E. coli (PDB ID 2PL1) in presence of beryllium fluoride ion (gold) and magnesium ion. Residues are shown as cylinder model. The beryllium fluoride (grey) and magnesium (blue) ions of the receiver domain response regulators in active state are shown as ball‐and‐stick model. D., E., and F. Superposition of RcsB C‐terminal DNA‐binding (lilac) and N‐terminal receiver domain (dark purple) with ChrA (grey), VraR (green), and NarL (gold) regulator. The average RMSD between superimposed RcsB C‐terminal and N‐terminal domains and homologous domains of full‐length FixJ/NarL regulators are ∼ 1.8 and 1.6 Å, respectively. Structures were superimposed in program CCP4mg.
