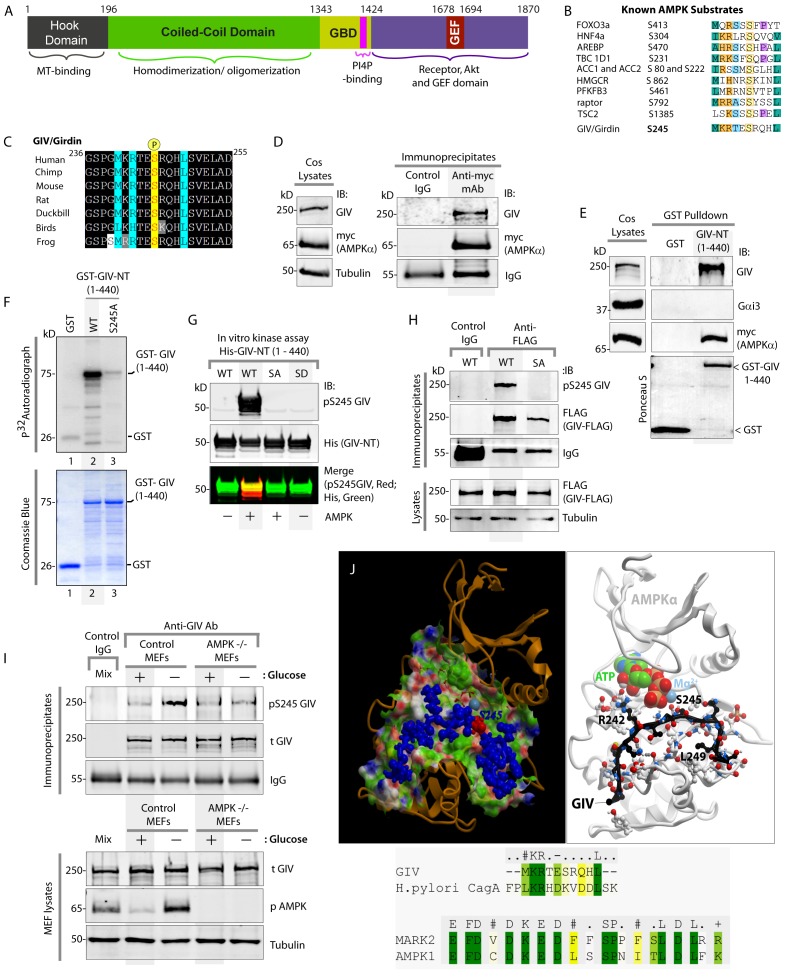
Figure 1. AMPK binds and phosphorylates GIV at Ser (S) 245.
(A) Schematic showing the functional modules of the multimodular signal transducer GIV. From the N- to the C-terminus the domains are-- a Hook-domain (grey) which binds microtubules (Simpson et al., 2005); a long coiled-coil domain (green) assists in homo/oligomerization (Enomoto et al., 2005); a Gα-binding domain (GBD; yellow) which constitutively binds Gαi/s proteins (Le-Niculescu et al., 2005); a PI(4)P-binding motif (pink) which enables GIV to bind PI4P-enriched membranes at the Golgi and the PM (Enomoto et al., 2005); an evolutionarily conserved GEF motif (red) which binds and activates Gαi (Garcia-Marcos et al., 2009) and inactivates Gαs (Gupta et al., 2016), and releases ‘free’ Gβγ from both. The C-terminal ~200 aa of GIV (purple) also has key domains that enable GIV to bind and remodel actin (Enomoto et al., 2005), bind and enhance phosphorylation of Akt (Anai et al., 2005; Enomoto et al., 2005), bind ligand-activated RTKs (Ghosh et al., 2010; Lin et al., 2014), and bind and activate Class 1 PI3-Kinases (Lin et al., 2011). (B) Consensus phosphorylation site for previously identified substrates of AMPK are aligned with the putative AMPK substrate site in human GIV. Conserved residues are highlighted with colors. (C) The sequence encompassing the putative AMPK substrate motif was aligned among various species using ClustalW. Conserved residues are shaded in black and similar residues in gray. The consensus residues within the sequence are highlighted in blue. The residue, Ser(S)245 which was predicted to be phosphorylated by AMPK is highlighted in yellow. (D) Immunoprecipitations were carried out on lysates of Cos7 cells expressing myc-AMPKα2 using anti-myc mAb. Immune complexes were analyzed for endogenous GIV and myc (AMPKα2) by immunoblotting (IB). (E) Lysates of Cos7 cells expressing myc-AMPKα2 were used as a source of AMPK in pulldown assays with bacterially expressed GST or GST-GIV-NT (aa 1–440; which includes S245) immobilized on glutathione beads. Bound proteins were analyzed for myc (AMPKα), Gαi3 (negative control; because this G protein binds GIV's C-terminus, not N-terminus) and endogenous GIV (positive control; because GIV homo-oligomerizes via its NT) by immunoblotting (IB). (F) In vitro kinase assays were carried out using recombinant AMPK heterotrimers (α2/β/γ) and bacterially expressed and purified GST-GIV-NT (1–440) proteins or GST alone (negative control) and γ -32P [ATP]. Phosphoproteins were analyzed by SDS-PAGE followed by autoradiography (top). Equal loading of substrate proteins was confirmed by staining the gel with Coomassie blue (bottom). AMPK phosphorylated GST-GIV-NT WT, but not the non-phosphorylatable SA mutant or GST alone. (G) Biochemical validation of a phosphospecific rabbit polyclonal antibody which detects GIV exclusively when it is phosphorylated at S245. In vitro kinase assays were carried out as described above and incubated in the presence of cold ATP. Phosphoproteins were analyzed for pS245-GIV and His (GIV-NT) by immunoblotting (IB). (H) In cellulo kinase assays were carried out in Cos7 cells co-expressing GIV-FLAG (WT or SA mutant) and myc-AMPKα constructs, and stimulating AMPK by glucose deprivation for 6 hr prior to lysis. GIV was immunoprecipitated from these lysates using anti-FLAG mAb and analyzed for phosphorylation of GIV at S245 by immunoblotting (IB) with anti-pS245-GIV and FLAG (GIV-Flag). GIV-WT, but not GIV-SA is phosphorylated at S245 in cells responding to energetic stress. (I) AMPK-/- and control MEFs were subjected or not to energetic stress by exposing them to growth conditions with (+) or without (-) glucose for 4 hr prior to lysis. GIV immunoprecipitated from equal aliquots of lysates (lower panel) were analyzed for total (t) and phosphorylated (pS245) GIV by immunoblotting. Representative blots are shown (n = 3). (J) Top: Homology model of GIV-bound AMPKα generated using the solved crystal structure of constitutively active Par1-MARK2 (a member of the AMPK family of kinases) in complex with the CagA protein encoded by pathologic strains of Helicobacter pylori [PDB: 3IEC (Nesic et al., 2010)] as template. Bottom: The target:template alignment is shown for the GIV peptide (with H. pylori CagA protein) and AMPK (with MAPK2, partial alignment of binding site residues).