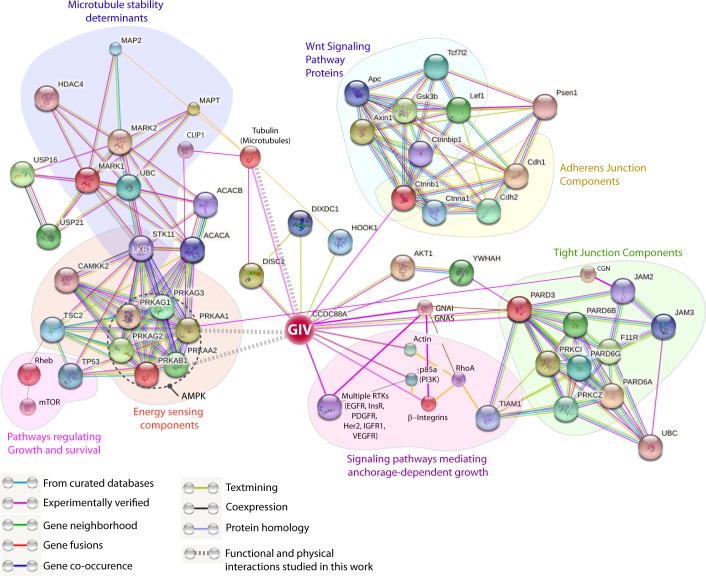
Figure 10. GIV (CCDC88A) functions at the nexus of a variety of cellular pathways that regulate and respond to changes in cell-cell junctions.
A protein-protein interaction network (see Materials and methods) shows all known and predicted protein-protein interactions (relevant to the current work), both functional and directly physical in nature, that stem from computational prediction, from knowledge transfer between organisms, and from interactions reported in other (primary) databases. The clusters of functional associations shown here are meant to be specific and meaningful, i.e. proteins jointly contribute to a shared function; this does not necessarily mean they are physically binding each other. The interaction network shows that GIV (CCDC88A) functions as a key polarity-determinant scaffold which links major components that sense cellular energy (STK11, LKB1; PRKA, AMPK) and determine cellular growth and survival responses (on the left) and signaling pathways, e.g., Wnt (top; light blue) and growth factor and integrin pathways (purple; bottom) that dictate anchorage-dependent vs independent growth in normal and transformed cells, respectively, to cell-cell junctions (both tight and adherens; on the right). The microtubule stability determinants (upper left; dark blue) are likely to synergize with the AMPK-GIV axis for protecting junctions against stress-induced collapse. Two previously defined functional interactions of PRKA (AMPK) studied in different contexts stand out as key links: (1) Phosphorylation of the microtubule plus end protein CLIP-170 (CLIP1; deep blue) by AMPK enhances the speed of microtubule polymerization (Nakano et al., 2010); and (2) Phosphorylation of the TJ- protein cingulin (CGN; green) by AMPK increases the association of plus ends of microtubules with TJs (Yano et al., 2013). Key at the lower left corner indicates how each functional interaction within this network was color-coded.