Figure 1.
Purification, Functional Characterization, and Expression of Immune Checkpoints in Tumor Infiltrating Cells
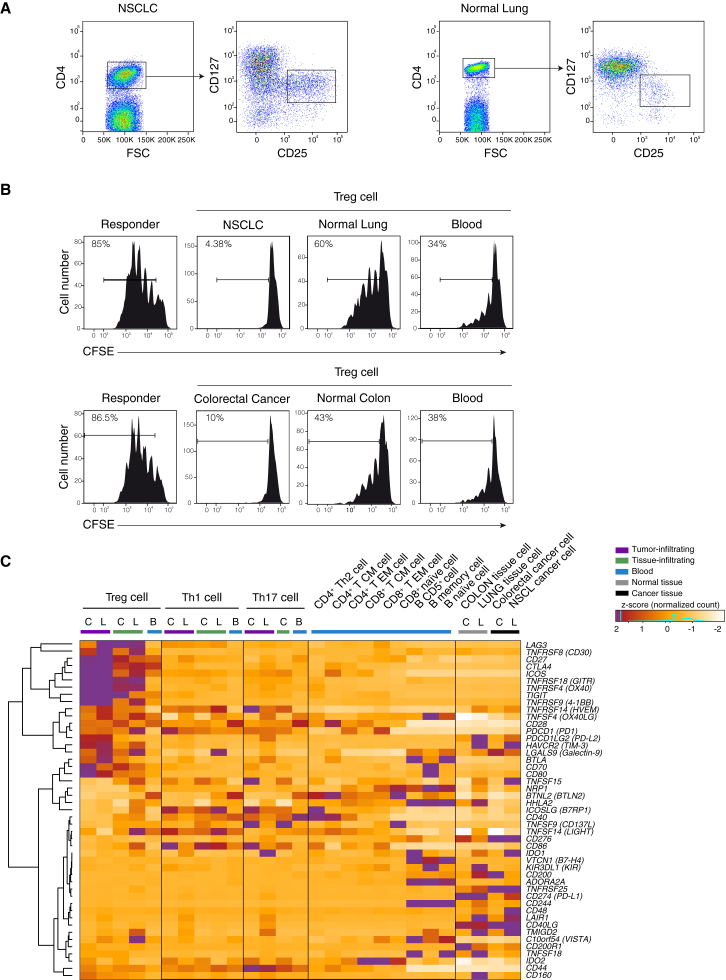
(A) Representation of the sorting strategy of Treg cells infiltrating tumor or normal tissue.
(B) Representative flow cytometry plots showing suppressive activity of Treg cells isolated from tumor (NSCLC or CRC), normal tissue and blood of the same patient. 4 × 105 carboxyfluorescein diacetate succinimidyl ester (CFSE)-labeled CD4+ naive T cells from healthy donors were cocultured with an equal number of Treg cells for 4 days with a CD3-specific mAb and CD1c+CD11c+ dendritic cells. Percentage of proliferating cells is indicated. Data are representative of three independent experiments.
(C) Z-score normalized RNA-seq expression values of immune checkpoints genes are represented as a heatmap. Cell populations are reported as a color code in the upper part of the graph, while gene names have been assigned to heatmap rows. Hierarchical clustering results are shown as a dendrogram drawn on the left side of the matrix. Colon tissues are indicated as C, lung tissues as L, and peripheral blood as B.
See also Figure S1.