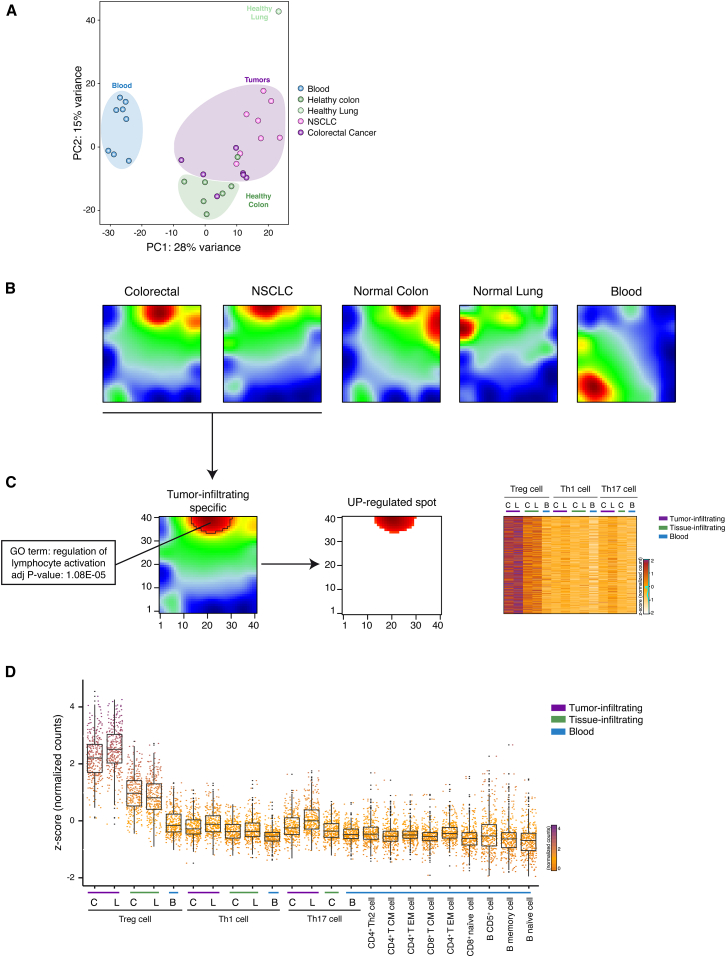
Figure 2.
SOM Analysis Identifies Co-regulated Genes in Tumor Infiltrating Treg Cells
(A) PCA has been performed on rlog-normalized (DESeq2) counts for all T regulatory cell RNA-seq samples (36 samples from 18 individuals).
(B) Self-organizing maps analysis has been performed on the RNA-seq dataset comprising Treg, Th1, and Th17 cell subsets. Bidimensional SOM profiles are reported for Treg cells.
(C) Group-centered analysis for the identification of upregulated spot (FDR < 0.1) in Treg cells infiltrating both NSCLC and CRC is described as 2D heatmap. Heatmap representing Z-score normalized expression values of genes selected from the upregulated spot is shown on the right side of the figure. Top enriched GO term (DAVID) for genes assigned to upregulated spot is reported with the corresponding significance p value. Colon tissues are indicated as C, lung tissues as L, and peripheral blood as B.
(D) Z-score normalized expression values of genes that are preferentially expressed in tumor-infiltrating Treg cells (Wilcoxon Mann Whitney test p < 2.2 × 10–16) over the listed cell subsets are represented as boxed plots. Colon tissues are indicated as C, lung tissues as L, and peripheral blood as B.
See also Figure S2.