Figure S5.
Details of Pre-accommodation Architectures, Related to Figure 5
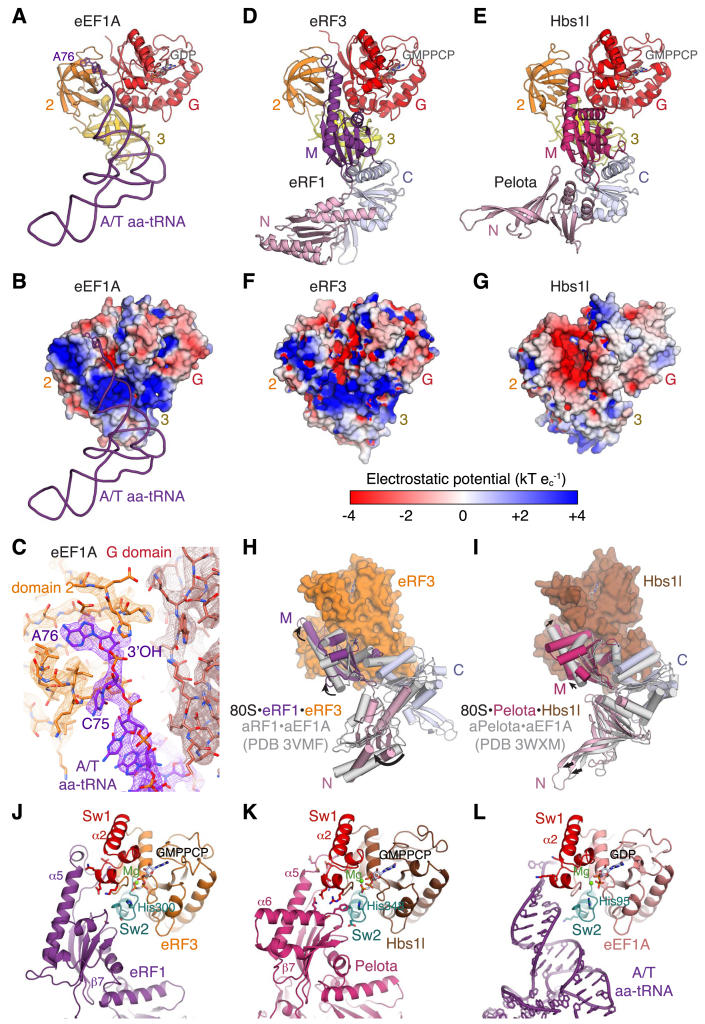
(A) The acceptor stem of aa-tRNA (purple) binds in a cleft between the G domain (red) and domains 2 (orange) and 3 (yellow) of eEF1A.
(B) Surface model of eEF1A colored by electrostatic potential (same view as panel A).
(C) EM map density contoured at 7σ and models of the interactions between the 3′ end of aa-tRNA (purple) and domain 2 (orange) and G domain (red) of eEF1A.
(D and E) The M domains of (D) eRF1 and (E) Pelota bind their respective GTPase partners in a cleft analogous to where aa-tRNA binds eEF1A. Structures are aligned as in panel (A).
(F and G) Surface model colored by electrostatic potential of (F) eRF3, and (G) Hbs1l.
(H and I) Superposition of (H) the crystal structure of aRF1⋅aEF1A⋅GTP (gray) on ribosome-bound eRF1⋅eRF3⋅GMPPCP or of (I) the crystal structure of aPelota⋅aEF1A⋅GTP (gray) on ribosome-bound Pelota⋅Hbs1l⋅GMPPCP via domains 2 and 3 of the GTPase. Upon ribosome binding, the N domain of the decoding factor is reoriented, while the M domain forms additional contacts with the G domain of the GTPase.
(J and K) Interactions between the M domains of (J) eRF1 or of (K) Pelota with the G domain of the respective GTPase. The β7-α5 loop, which harbors the GGQ motif of eRF1, makes interactions with the Switch 1 (Sw1, red) motif, and additional interactions are formed with the Switch 2 (Sw2, teal) motif harboring the catalytic histidine.
(L) The backbone and CCA end of A/T aa-tRNA also interacts with catalytically important motifs of the G domain of eEF1A.