Figure 5.
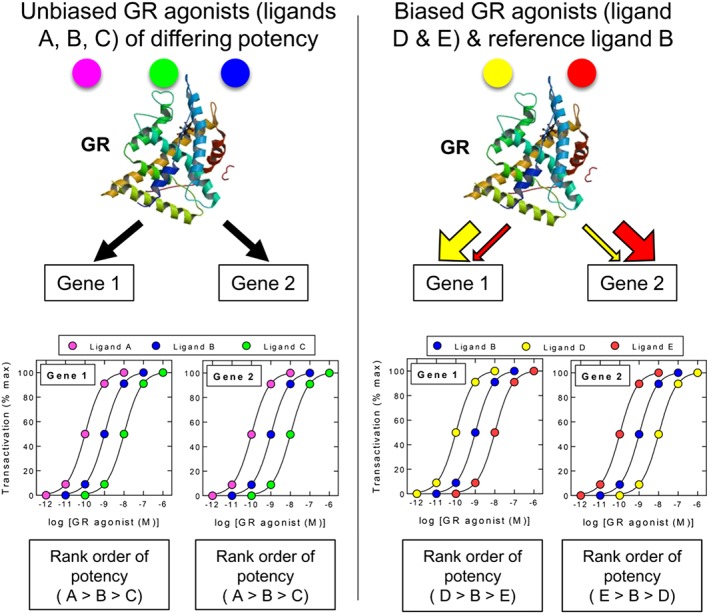
Biased agonism at GR. The possibility that nuclear hormone receptors such as GR can adopt different active conformations in a ligand‐dependent manner could provide a means to promote gene expression bias in a given tissue. According to this hypothetical model, unbiased ligands (A, B and C) generate a GR conformation(s) that promotes the expression of the GC transcriptome (shown here as Gene 1 and Gene 2) with the same rank order of potency where, in the figure, ligand A is more potent than ligand B, which is more potent that ligand C on all genes. In contrast, biased agonists (D and E) lead to the stabilization of different GR conformers that more favourably promote the transcription of one gene (or gene population) over another relative to the reference ligand B. Gene expression bias could result from either a change in the way GR interacts with essential co‐factors and/or its ability to bind DNA and effect transcription. In each case, this will be dictated by the promoter context of each target gene. The rank order of agonist potency can therefore vary in a gene‐dependent manner as depicted by the concentration–response curves describing ligands D and E relative to the reference ligand B. Figure generated using the GR ribbon diagram found at http://www.rcsb.org/pdb/explore/explore.do?pdbId=1P93
