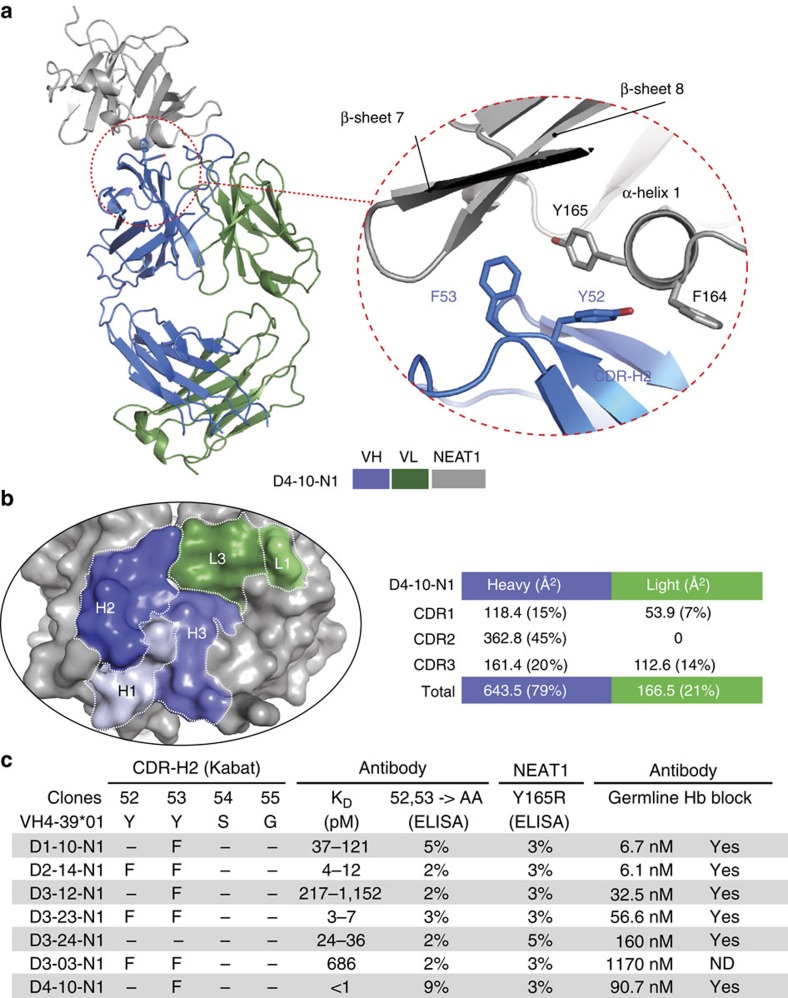
Figure 4. Germline-encoded binding of IGHV4-39 to the NEAT1 domain of IsdB.
(a) Crystal structure of an IGHV4-39-derived Fab (D4-10-N1) in complex with NEAT1. The two aromatic resides (Y52 and F53) in CDR-H2 interact with the α-helix1 of NEAT1 which is normally involved in binding haemoglobin. IGHV4-39 CDR-H2 F53 of Fab D4-10-N1 protrudes into a hydrophobic pocket of NEAT1, which is structurally homologous to the heme binding pocket of NEAT2. Crystallization was facilitated by the use of a sandwiching Fab from an antibody (D3–19) that binds NEAT1 at a non-overlapping epitope (bin H). For clarity, the sandwiching Fab is removed from the figure but is included in the Supplementary Data (Supplementary Fig. 10c). (b) CDR-H2 dominates the interaction in terms of BSA. Structural analysis shows that 79% of the BSA is attributed to the heavy chain, and 21% to the light chain. The CDR-H2 contributes about 45% of total BSA. (c) Mutational analysis confirms the structural data and demonstrates that all IGHV derived antibodies in this set bind NEAT1 with a similar mechanism. The KD for all antibodies in this set was determined by SPR-based biosensor binding analysis to recombinant full-length IsdB at 37 °C (KD range, n≥2). The binding of antibody variants at positions 52 and 53 of CDR-H2 to wild type IsdB and the binding of antibodies to NEAT1 variant Y165R (α-helix 1) were evaluated by ELISA (percentage binding relative to binding between original isolated antibodies and wild type IsdB, one representative set of results out of three independent experiments is shown). Every clone was reverted to VH germline sequence and tested for binding to NEAT1 by biosensor analysis and ability to block haemoglobin binding. ND stands for not determined.