Figure 7.
An N-terminal Low-Complexity Region in Mmi1 Is Critical for Stable Interaction with Ccr4-Not and Stimulation of Deadenylation
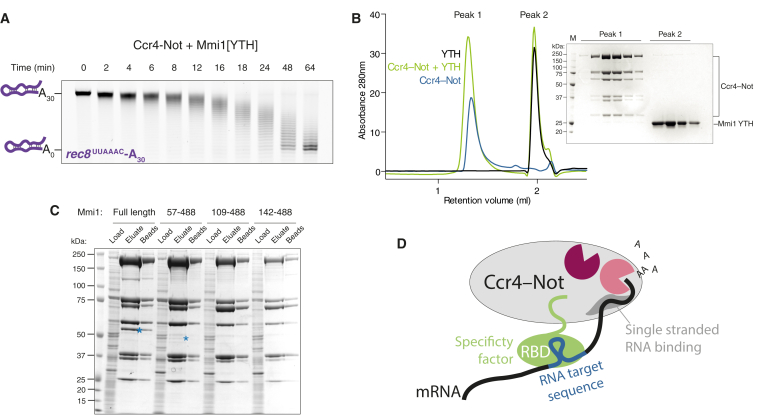
(A) The Mmi1 YTH domain does not stimulate deadenylation in trans. Deadenylation assay was performed on the rec8-A30 substrate with recombinant Ccr4-Not and an equimolar quantity of Mmi1 YTH RNA-binding domain.
(B) Analytical size exclusion chromatogram with SDS-PAGE of peak fractions demonstrating that Ccr4-Not and the Mmi1 YTH domain construct do not co-migrate. Ccr4-Not alone (blue) and the Mmi1 YTH domain alone (black) are also shown. M, molecular weight marker.
(C) The first 56 N-terminal amino acids of Mmi1 are critical for stable complex formation with Ccr4-Not, as shown in SDS-PAGE analysis of pull-downs of proteins expressed from recombinant baculoviruses containing Mmi1 N-terminal deletions. Blue asterisks denote the Mmi1 protein where it can be observed.
(D) Model for targeted deadenylation by Ccr4-Not. The low-complexity region of a specificity factor (green, e.g., Mmi1) is required for interaction with Ccr4-Not (gray, shown with two deadenylases in pink), whereas the RNA-binding domain of a specificity factor (e.g., the C-terminal YTH domain of Mmi1) binds target mRNAs (e.g., DSR sequence, blue), resulting in mRNA deadenylation. The Ccr4-Not complex may also contain an intrinsic single-stranded-RNA-binding activity.