Abstract
The AlkB gene that protects E.coli against methylation damage to DNA was identified more than 3 decades ago. 20 years later, the AlkB protein was shown to catalyze repair of methylated DNA base lesions by oxidative demethylation. Two human AlkB homologs were characterized with similar DNA repair activities and seven additional human AlkB homologs were identified based on sequence homology. All these dioxygenases, ALKBH1–8 and FTO, contain a conserved α-ketoglutarate/iron-dependent domain for methyl modifications and de-modifications. Well-designed research over the last 10 years has identified unforeseen substrate heterogeneity for the AlkB homologs, including novel reversible methyl modifications in RNA. The discoveries of RNA demethylation catalyzed by AlkB family enzymes initiated a new realm of gene expression regulation, although the understanding of precise endogenous activities and roles of these RNA demethylases are still undeveloped. It is worth mentioning that the AlkB mechanism and use of α-ketoglutarate have also emerged to be essential for many enzymes in epigenetic reprogramming that modify and de-modify methylated bases in DNA and methylated amino acids in histones.
Keywords: AlkB homologs, DNA repair, RNA metabolisms, Oxidative demethylation, YTH domain proteins
1. Introduction
In 1983, Sekiguchi and coworkers isolated seven mutants of E. coli that were sensitive to methylating agent but not to UV light [1]. Most of these mutations were mapped to the region of the E. coli genome encoding the well known AlkA DNA glycosylase. One mutation was novel and the gene responsible was named AlkB. Despite many studies, no function was assigned to the E. coli AlkB gene for 2 decades. Yet, an important clue to the activity of the AlkB protein was reported in 2000; it was shown that the reactivation of methylated single-stranded DNA phages was considerably reduced in E. coli lacking the AlkB gene [2]. One year later, Aravind and Koonin [3] employed sequence profiling analysis to show that AlkB and other enzymes define a novel family of the 2OG-Fe(II) oxygenase superfamily. They conclude that the AlkB protein catalyzes oxidative detoxification of alkylated bases and further suggest that the more distant homologs of AlkB detected in eukaryotes and in plant RNA viruses might be involved in RNA demethylation. These bioinformatics analysis were soon to be verified experimentally for the E. coli AlkB and two human homolgos, ALKBH2 and ALKBH3 [4–7]. A year later, five more human homologs of the E.coli AlkB enzyme are identified, ALKBH4–8 [8]. They hypothesize that these homologs could be back-up enzymes for the DNA repair activities of Alkbh2 and 3 or may code for novel DNA or RNA repair activities. With the addition of the FTO (fat mass and obesity-associated) protein the mammalian family of AlkB proteins now consists of 9 members (Fig. 1). Recent research has identified remarkable diverse substrate specificities for the AlkB homologs. Yet, the activities of some homologs are still hard to pin down. In this review, we focus on the AlkB homologs acting on RNA.
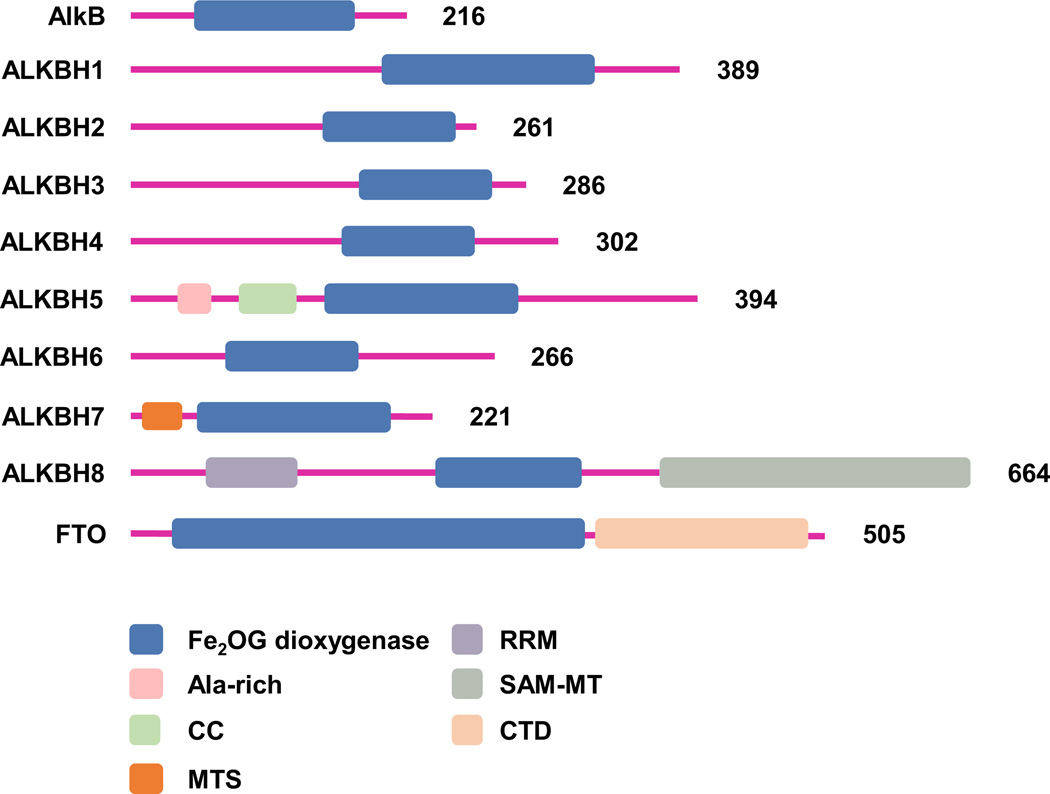
Fig. 1.
Domain architecture of the Escherichia coli (E. coli) AlkB and the nine mammalian AlkB homologs. UniProtKB accession number for E. coli AlkB (P05050); and human ALKBH1(Q5XKL0), ALKBH2(Q6NS38), ALKBH3(Q96Q83), ALKBH4(Q9NXW9), ALKBH5(Q6P6C2), ALKBH6(Q3KRA9), ALKBH7(Q9BT30), ALKBH8(Q96BT7) and FTO(Q9C0B1). Fe2OG dioxygenase (Fe(II) and α-ketoglutarate dependent dioxygenase domain); CC (Coiled Coil), MTS (Mitochondrial Targeting Signal), Ala-rich (Region with Ala stretch); RRM (RNA Recognition Motif), SAM-MT (S-adenosylmethionine dependent Methyltransferase domain), and CTD (C-Termina Domain).
2. Past and present of AlkB mediated RNA repair
Researchers have speculated on the functional roles of AlkB family proteins on RNA modifications. In 2003, reviewing the novel finding of E. coli and human AlkBs in mediating DNA, and possible RNA, repair by oxidative demethylation, Begley and Samson suggested that the “RNA repair” activities of AlkB and ALKBH3 might have evolved to regulate biological RNA methylation [9]. In 2010, we proposed that post-transcriptional RNA modifications can be dynamic and may play additional roles in gene expression regulation in a grand challenge commentary [10]. The AlkB-mediated demethylation was specifically proposed as a potential pathway for RNA demodification. In recent years these assumption has been confirmed in several important studies. However, the RNA repair activities of AlkB and its human homolog’s have not been con-firmed in vivo, although recombinant AlkB from E. coli and ALKBH3 from mammals efficiently demethylate 1 mA and 3mC in RNA. In an early study, these hydroxylases were shown to restore the biological function of chemically methylated mRNAs and tRNAs [11]. As the relevant modifications, 1mA and 3mC, also are naturally occurring modifications in some RNAs, introduced enzymatically, they speculated that a low level of AlkB-mediated removal of enzymatically introduced methyl modifications may be accepted, due to rapid replenishment by the methyltransferases. This raises the same question as for the 5-hydroxymethyluracil (5hmU) modification in DNA. In a recent study [12], TET is shown to directly produce 5hmU in mouse embryonic stem cell DNA by oxidizing thymine (T). Thus, 5hmU in DNA is introduced by TET enzyme(s) and by reactive oxygen species and at present there is no indications that the SMUG glycosylase that recognize and excise 5hmU has a specificity or partner proteins that can aid in distinguishing between ROS and TET induced 5hmU. Thus, the historical distinction on chemically introduced damaged bases in DNA and RNA as opposed to enzymatically introduced regulatory bases in DNA and RNA might have to be seen in a new light.
3. Regulatory DNA modifications
Known regulatory modifications in mammalian genomic DNA have for long been restricted to the methylation of the 5-position of cytosine (5mC). In recent years, the further hydroxylation of 5mC to 5-hydroxymethylCytosine (5hmC), 5-formylCytosine (5fC) and 5-carboxylCytosine (5caC) by the AlkB like TET enzymes have added complexity to the understanding of modified cytosines in DNA. It is very interesting to notice that the final removal of these cyto-sine modifications is carried out by DNA glycosylases [13]. Thus, the processing of nearby cytosine modifications in opposing DNA strands, like clustered oxidative DNA damage, by the base excision repair (BER) enzymes must be coordinated to avoid producing double-strand DNA breaks (DSBs) [14]. In a recent study [15] it is shown that TET1-TDG-BER dependent active DNA demethylation is highly coordinated to avoid double strand breaks in DNA. Very recently, the repertoire of regulatory methylated bases in mammalian genomic DNA was expanded to also include methylation of the 6-position of adenine (6mA) and the possible dynamic nature of this modifications could well indicate a role for an AlkB-like enzyme for its reversal [16–18]. For this review, it is worth mentioning that histone demethylation by hydroxylation also share homology to AlkB demethylation. The similarity of removing a methyl group from Adenine (1mA) and methylated-lysine led Shi and co-workers to discover the first histone demethylase [19], and Zhang and co-workers to look for a similar mechanism for histone lysine demethylation and they identified JHDM1 (JmjC domain-containing histone demethylase 1) following purification through six chromatography columns [20].
While the number of regulatory bases in DNA is relatively limited, there are a wide variety of regulatory bases in RNA that are often quite abundant. The known biochemical processes involved in generating and removing methylated bases from DNA and RNA are remarkably similar and here we will focus on the recent identification of the reversible status of RNA modifications and the role of the mammalian AlkB enzymes.
4. ALKBH8 mediated tRNA modifications
Amongst the nine AlkB homologs, ALKBH8 is unique by containing one additional annotated protein domain. The activity of this C-terminal methyltransferase, methylation of certain wobble uridines in some tRNAs, was elucidated due to its homology to the S. cerevisiae tRNA methyltransferase, Trm9 [21,22]. The AlkB domain of ALKBH8 was later shown to catalyze the hydroxylation of 5-methoxycarbonylmethyluridine (mcm5U) at the wobble position of certain tRNAs [23,24]. This generate, unlike the other known AlkB enzymes that produces an unstable hydroxymethylated base that is spontaneously demethylated, a new stable hydroxylated base comparable to the hydroxylation of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) in genomic DNA [23,24].
5. Reversible methylations of mRNA
The research of RNA modifications entered a new era in 2011 with the identification of the reversible nature of the N6-methyladenine modifications by the Fe(II)- and α-ketoglutarate-dependent fat mass and obesity-associated (FTO) protein [25] (Fig. 2). Soon after that, a second Fe(II)- and α-ketoglutarate-dependent hydroxylase, ALKBH5, was also shown to demethylate m6A in RNA [26]. m6A is a very common internal modification in mRNA and non-coding RNAs. It was also shown that two additional modifications, 6-hydroxymethyladenosine (hm6A) and 6-formyladenosine (f6A), exist in mammalian messenger RNA and that the FTO protein can further oxidize m6A to make hm6A and f6A [27]. Following the discovery of FTO, two laboratories developed sequencing methods to profile m6A in the mammalian transcriptomes [28,29]. In three recent studies [30–32] m6A-CLIP (cross-linking and immunoprecipitation) followed by reverse transcription was used to map m6A at high-resolution throughout the transcriptome. The mutational signature induced by the reverse transcriptase allows for potential single-base resolution mapping of m6A throughout the transcriptome. Although these methods are not suitable for use in low input materials, they allowed further insight on, for example, the location of m6A and its possible role in regulating alternative polyA choice.
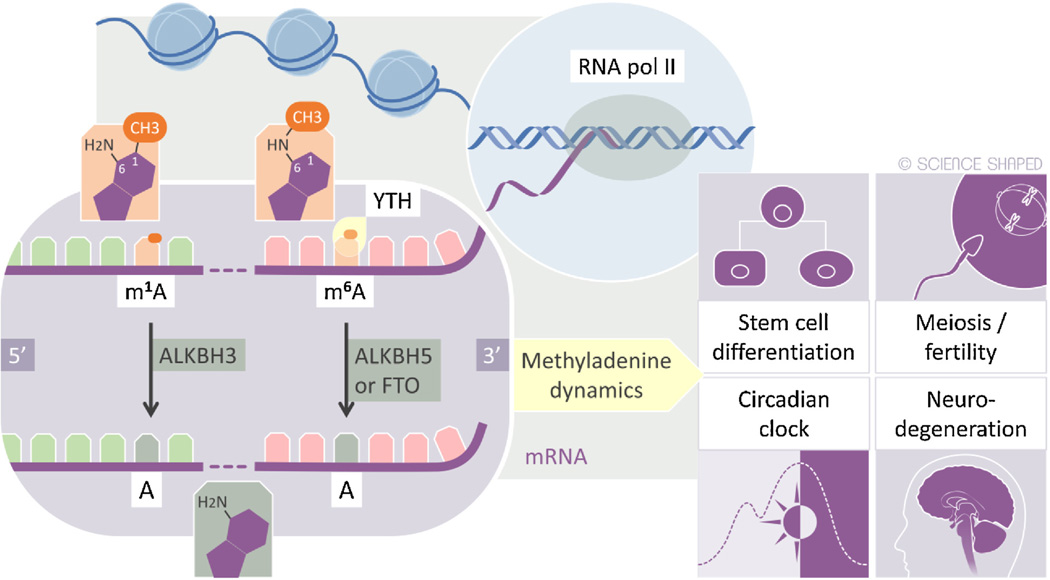
Fig. 2.
Overview of the distinct pattern of 1-methyladenine (m1A) and 6-methyladenine (m6A) dynamics in mammalian mRNA. Transcriptome mapping of m1A in mRNA was done very recently so the consequences of methylation dynamics indicated is for m6A. All other details in the main text.
High-resolution mapping allowed further insight on the location of m6A and suggest a role for m6A in regulating alternative polyA choice [31]. Similarly, 5-hydroxymethylcytosine (5hmC), a modifications that recently have attracted much attention due to its presence in genomic DNA, also occurs in RNA and was recently shown to be introduced by the TET enzyme and mark polyadenylated RNAs in Drosophila [33]. Transcriptome mapping of cells and organisms lacking ALKBH5 and/or FTO has not yet been completed. Thus, no information on the specificity of the methyltransferases METTL3 and METTL14 [34] or the demethylases ALKBH5 and FTO exist and data published on the sequence specificity of m6A in the mammalian genome so far thus only portray the combined effect of m6A deposition by methyltransferases and its removal by demethylases. The selectivity of the methylation deposition and removal remain unanswered questions.
The discovery and characterization of reader proteins that recognize m6A have significantly advanced functional understanding of this modification in gene expression regulation. The YTH family proteins selectively recognize methylated RNA and affect metabolism and processing of the target mRNAs [28,35,36]. The methylation can promote translation, mediate mRNA decay, and affects localization and splicing. Upon heat shock, YTHDF2 can preserve m6A methylation of stress-induced transcripts [37]. The m6A methylation also occurs on lncRNA and pri-miRNA. It serves as a structural switch to tune RNA secondary structure and affects binding by RNA-binding proteins [38]. The methylation on pri-miRNA can be recognized by HNRNPA2B1 to accelerate miRNA maturation [39].
More recently, two papers portray 1-methylAdenine (m1A) in eukaryotic mRNA [40,41], and describe a potential role for ALKBH3 for its reversible and dynamic nature. m1A modifications in mRNA are less frequent than m6A, 10-fold less in cell lines but 3-fold less in tissues, and can occur on a few thousands of mRNA transcripts in humans. Unlike m6A, m1A appears around start codon and strongly correlates with protein production. The unique features of m1A indicate a role in translation of methylated mRNA. The methyltransferase introducing the m1A modification is not yet identified.
6. The biology of reversible modifications in RNA
The reversible nature of some RNA modifications is a very recent discovery and the biological consequences of aberrant dynamics of m6A (and m1A) have therefore just now become a potential topic of study. Based on mutant analysis in model organisms, it seems clear that m6A modifications in mRNA have unique roles in meiosis [26,42–44] (Fig. 2). The precise role of individual mRNAs may now be studied due to the ability to map m6A sites that are dynamically regulated during meiosis. The m6A modification in mRNA also has crucial roles for embryonic stem cells and depletion of the METTL3 m6A methyltransferase results in impaired differentiation and restricted lineage priming [45,46]. It is also shown that m6A in mRNA is associated with the timing of circadian periods [47].
Initially, the extraordinary interest in FTO was sparked by its association with obesity. However, the relevant polymorphic variants in the introns of the FTO gene does not seem to affect FTO expression or activity and it is more likely that these polymorphisms have an effect, as a long-range promoter, for the downstream IRX3 gene [48]. Less attention has been credited to the study that revealed that an inactivating FTO mutation is responsible for an autosomal-recessive lethal syndrome which is suggestive of a crucial role of FTO in global gene expression regulation [49]. The in vivo relevant RNAs serving as substrate for FTO is still unknown and mutant cells and organisms should be used for its identification.
7. Future studies
The dynamically regulated m6A and m1A bases in RNA are chemically stable and recent data indicate that they carry out their function by interacting with reader proteins. The distinct and highly conserved positioning of these modifications further argues for such a role. Yet, m6A and m1A could also affect hydrogen binding in certain RNA structures since the positions methylated are involved in “Watson-crick” base pairing. In order to advance our understanding of RNA methabolisms by reversible base modifications, focus could be on:
The development of quantitative sequencing methods for m6A with less input materials (current methods require millions of cells). Highly accurate sequencing results could allow site-directed mutagenesis studies to get insight on individual mRNA transcripts that are dynamically regulated by environmental stress or through periodic regulations like the cell cycle and the circadian rhythm.
Biochemical and cellular characterization of the writer complex, additional readers, and existing and potentially new erasers, including structural characterizations.
The extensive post-transcriptional gene expression regulation through RNA methylation in various biological processes, as well as their impacts on various human diseases.
Posttranslational events that impact RNA methylation writers, readers and erasers to reveal how selective writing, reading and erasing are achieved.
More than 100 post-transcriptional base modifications have been identified in various RNA-species. Thus, it is very likely that many of these are dynamic or even reversible. The impacts of dynamic and potentially reversible modifications on other RNA species such as tRNA and rRNA.
The activities of ALKBH6 and ALKBH7 remains a mystery. ALKBH7 is the only AlkB homolog containing a conserved mitochondrial signal and is involved in programmed necrosis and fat metabolism [50,51]. The exact role and underlying mechanism of ALKBH4 still need further studies [52].
Methylated bases occur frequently in a variety of RNA species and the specificity of demethylases is not well characterized regarding the possibility that other factors restrict or initiate substrate recognition. Also, individual enzymes might have several relevant substrates, both regarding the modification itself and the type of RNA molecule.
Acknowledgments
This work was supported by the Norwegian Research Council and the Research Program of the EEA/Norway Grants (grant number POL/NOR/196258/2013) and National Institutes of Health NIGMSGM071440 (C.H.). C.H. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Kataoka H, Yamamoto Y, Sekiguchi M. A new gene (alkB) of Escherichia coli that controls sensitivity to methyl methane sulfonate. J. Bacteriol. 1983;153:1301–1307. doi: 10.1128/jb.153.3.1301-1307.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dinglay S, Trewick SC, Lindahl T, Sedgwick B. Defective processing of methylated single-stranded DNA by E.coli AlkB mutants. Genes Dev. 2000 Aug;14(16):2097–2105. [PMC free article] [PubMed] [Google Scholar]
- 3.Aravind L, Koonin EV. The DNA-repair protein AlkB, EGL-9, and leprecan define new families of 2-oxoglutarate- and iron-dependent dioxygenases. Genome Biol. 2001;2(3) doi: 10.1186/gb-2001-2-3-research0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trewick SC, Henshaw TF, Hausinger RP, Lindahl T, Sedgwick B. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature. 2002 Sep;419(6903):174–178. doi: 10.1038/nature00908. [DOI] [PubMed] [Google Scholar]
- 5.Falnes PO, Johansen RF, Seeberg E. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature. 2002 Sep;419(6903):178–182. doi: 10.1038/nature01048. [DOI] [PubMed] [Google Scholar]
- 6.Aas PA, Otterlei M, Falnes PO, Vagbo CB, Skorpen F, Akbari M, et al. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature. 2003 Feb;421(6925):859–863. doi: 10.1038/nature01363. [DOI] [PubMed] [Google Scholar]
- 7.Duncan T, Trewick SC, Koivisto P, Bates PA, Lindahl T, Sedgwick B. Reversal of DNA alkylation damage by two human dioxygenases. Proc. Natl. Acad. Sci. U.S.A. 2002 Dec;99(26):16660–16665. doi: 10.1073/pnas.262589799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kurowski MA, Bhagwat AS, Papaj G, Bujnicki JM. Phylogenomic identification of five new human homologs of the DNA repair enzyme AlkB. BMC Genom. 2003 Dec;4(1):48. doi: 10.1186/1471-2164-4-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Begley TJ, Samson LD. AlkB mystery solved: oxidative demethylation of N1-methyladenine and N3-methylcytosine adducts by a direct reversal mechanism. Trends Biochem. Sci. 2003 Jan;28(1):2–5. doi: 10.1016/s0968-0004(02)00010-5. [DOI] [PubMed] [Google Scholar]
- 10.He C. Grand challenge commentary: RNA epigenetics? Nat. Chem. Biol. 2010 Dec;6(12):863–865. doi: 10.1038/nchembio.482. [DOI] [PubMed] [Google Scholar]
- 11.Ougland R, Zhang CM, Liiv A, Johansen RF, Seeberg E, Hou YM, et al. AlkB restores the biological function of mRNA and tRNA inactivated by chemical methylation. Mol. Cell. 2004 Oct;16(1):107–116. doi: 10.1016/j.molcel.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 12.Pfaffeneder T, Spada F, Wagner M, Brandmayr C, Laube SK, Eisen D, et al. Tet oxidizes thymine to 5-hydroxymethyluracil in mouse embryonic stem cell DNA. Nat. Chem. Biol. 2014 Jul;10(7):574–581. doi: 10.1038/nchembio.1532. [DOI] [PubMed] [Google Scholar]
- 13.He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011 Sep;333(6047):1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cannan WJ, Tsang BP, Wallace SS, Pederson DS. Nucleosomes suppress the formation of double-strand DNA breaks during attempted base excision repair of clustered oxidative damages. J. Biol. Chem. 2014 Jul;289(29):19881–19893. doi: 10.1074/jbc.M114.571588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alain Weber CK, Robertson Adam B, Kusnierczyk Anna, Vågbø Cathrine, Schuermann David, Klungland Arne, Schar Primo. Biochemical reconstitution of TET 1-TDG-BER dependent active DNA demethylation reveals a highly coordinated mechanism. Nat. Commun. 2016;7:10806. doi: 10.1038/ncomms10806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greer EL, Blanco MA, Gu L, Sendinc E, Liu J, Aristizabal-Corrales D, et al. DNA methylation on N6-adenine in C: elegans. Cell. 2015 May;161(4):868–878. doi: 10.1016/j.cell.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang G, Huang H, Liu D, Cheng Y, Liu X, Zhang W, et al. N6-methyladenine DNA modification in drosophila. Cell. 2015 May;161(4):893–906. doi: 10.1016/j.cell.2015.04.018. [DOI] [PubMed] [Google Scholar]
- 18.Fu Y, Luo GZ, Chen K, Deng X, Yu M, Han D, et al. N6-methyldeoxyadenosine marks active transcription start sites in chlamydomonas. Cell. 2015 May;161(4):879–892. doi: 10.1016/j.cell.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004 Dec;119(7):941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 20.Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006 Feb;439(7078):811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 21.Songe-Moller L, van den Born E, Leihne V, Vagbo CB, Kristoffersen T, Krokan HE, et al. Mammalian ALKBH8 possesses tRNA methyltransferase activity required for the biogenesis of multiple wobble uridine modifications implicated in translational decoding. Mol. Cell. Biol. 2010 Apr;30(7):1814–1827. doi: 10.1128/MCB.01602-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu D, Brophy JA, Chan CT, Atmore KA, Begley U, Paules RS, et al. Human AlkB homolog ABH8 Is a tRNA methyltransferase required for wobble uridine modification and DNA damage survival. Mol. Cell. Biol. 2010 May;30(10):2449–2459. doi: 10.1128/MCB.01604-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van den Born E, Vagbo CB, Songe-Moller L, Leihne V, Lien GF, Leszczynska G, et al. ALKBH8-mediated formation of a novel diastereomeric pair of wobble nucleosides in mammalian tRNA. Nat. Commun. 2011;2:172. doi: 10.1038/ncomms1173. [DOI] [PubMed] [Google Scholar]
- 24.Fu Y, Dai Q, Zhang W, Ren J, Pan T, He C. The AlkB domain of mammalian ABH8 catalyzes hydroxylation of 5-methoxycarbonylmethyluridine at the wobble position of tRNA. Angew. Chem. Int. Ed. Engl. 2010 Nov;49(47):8885–8888. doi: 10.1002/anie.201001242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011 Decenber;7(12):885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 2013 Jan;49(1):18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu Y, Jia G, Pang X, Wang RN, Wang X, Li CJ, et al. FTO-mediated formation of N6-hydroxymethyladenosine and N6-formyladenosine in mammalian RNA. Nat. Commun. 2013;4:1798. doi: 10.1038/ncomms2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012 May;485(7397):201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 29.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012 Jun;149(7):1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods. 2015 Aug;12(8):767–772. doi: 10.1038/nmeth.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ke S, Alemu EA, Mertens C, Gantman EC, Fak JJ, Mele A, et al. A majority of m6A residues are in the last exons, allowing the potential for 3’ UTR regulation. Genes Dev. 2015 Oct;29(19):2037–2053. doi: 10.1101/gad.269415.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen K, Luo GZ, He C. High-resolution mapping of N(6)-methyladenosine in transcriptome and genome using a photo-crosslinking-assisted strategy. Methods Enzymol. 2015;560:161–185. doi: 10.1016/bs.mie.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 33.Delatte B, Wang F, Ngoc LV, Collignon E, Bonvin E, Deplus R, et al. RNA biochemistry. Transcriptome-wide distribution and function of RNA hydroxymethylcytosine. Science. 2016 Jan;351(6270):282–285. doi: 10.1126/science.aac5253. [DOI] [PubMed] [Google Scholar]
- 34.Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014 Feb;10(2):93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014 Jan;505(7481):117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015 Jun;161(6):1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature. 2015 Oct;526(7574):591–594. doi: 10.1038/nature15377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015 Feb;518(7540):560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alarcon CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF. HNRNPA2B1, Is a mediator of m(6)A-dependent nuclear RNA processing events. Cell. 2015 Sep;162(6):1299–1308. doi: 10.1016/j.cell.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, Peer E, Kol N, Ben-Haim MS, et al. The dynamic N-methyladenosine methylome in eukaryotic messenger RNA. Nature. 2016 Feb;(10) doi: 10.1038/nature16998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li X, Xiong X, Wang K, Wang L, Shu X, Ma S, et al. Transcriptome-wide mapping reveals reversible and dynamic N-methyladenosine methylome. Nat. Chem. Biol. 2016 Feb;(10) doi: 10.1038/nchembio.2040. [DOI] [PubMed] [Google Scholar]
- 42.Clancy MJ, Shambaugh ME, Timpte CS, Bokar JA. Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6-methyladenosine in mRNA: a potential mechanism for the activity of the IME4 gene. Nucleic Acids Res. 2002 Oct;30(20):4509–4518. doi: 10.1093/nar/gkf573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhong S, Li H, Bodi Z, Button J, Vespa L, Herzog M, et al. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell. 2008 May;20(5):1278–1288. doi: 10.1105/tpc.108.058883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hongay CF, Orr-Weaver TL. Drosophila Inducer of MEiosis 4 (IME4) is required for Notch signaling during oogenesis. Proc. Natl. Acad. Sci. U.S.A. 2011 Sep;108(36):14855–14860. doi: 10.1073/pnas.1111577108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Batista PJ, Molinie B, Wang J, Qu K, Zhang J, Li L, et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014 Dec;4(6):707–719. doi: 10.1016/j.stem.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, et al. Stem cells m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science. 2015 Feb;347(6225):1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- 47.Fustin JM, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M, et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell. 2013 Nov;155(4):793–806. doi: 10.1016/j.cell.2013.10.026. [DOI] [PubMed] [Google Scholar]
- 48.Smemo S, Tena JJ, Kim KH, Gamazon ER, Sakabe NJ, Gomez-Marin C, et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014 Mar;507(7492):371–375. doi: 10.1038/nature13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boissel S, Reish O, Proulx K, Kawagoe-Takaki H, Sedgwick B, Yeo GS, et al. Loss-of-function mutation in the dioxygenase-encoding FTO gene causes severe growth retardation and multiple malformations. Am. J. Hum. Genet. 2009 Jul;85(1):106–111. doi: 10.1016/j.ajhg.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Solberg A, Robertson AB, Aronsen JM, Rognmo O, Sjaastad I, Wisloff U, et al. Deletion of mouse Alkbh7 leads to obesity. J. Mol. Cell Biol. 2013 Jun;5(3):194–203. doi: 10.1093/jmcb/mjt012. [DOI] [PubMed] [Google Scholar]
- 51.Fu D, Jordan JJ, Samson LD. Human ALKBH7 is required for alkylation and oxidation-induced programmed necrosis. Genes Dev. 2013 May;27(10):1089–1100. doi: 10.1101/gad.215533.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li MM, Nilsen A, Shi Y, Fusser M, Ding YH, Fu Y, et al. ALKBH4-dependent demethylation of actin regulates actomyosin dynamics. Nat. Commun. 2013;4:1832. doi: 10.1038/ncomms2863. [DOI] [PMC free article] [PubMed] [Google Scholar]