Fig. 2.
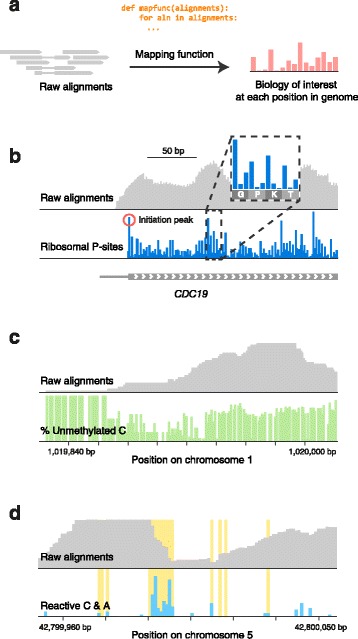
Mapping functions extract biological data from read alignments. a. Mapping functions use various properties of a read alignment to determine the genomic position(s) at which it should be counted. b. Mapping functions for ribosome profiling use alignment coordinates and lengths to estimate ribosome positions, revealing features of translation, like a peak of density at the start codon (red circle) and three-nucleotide periodicity of ribosomal translocation (inset). c. For bisulfite sequencing, the fraction of C-to-T transitions at each cytosine are mapped, revealing a CpG island. d. A mapping function for DMS-seq differentiates structured from unstructured regions of a selenocysteine insertion element in the 3′ UTR of human SEPP1. DMS reactivity (blue bars) matches A and C residues predicted to be unstructured (yellow)
