Figure 2.
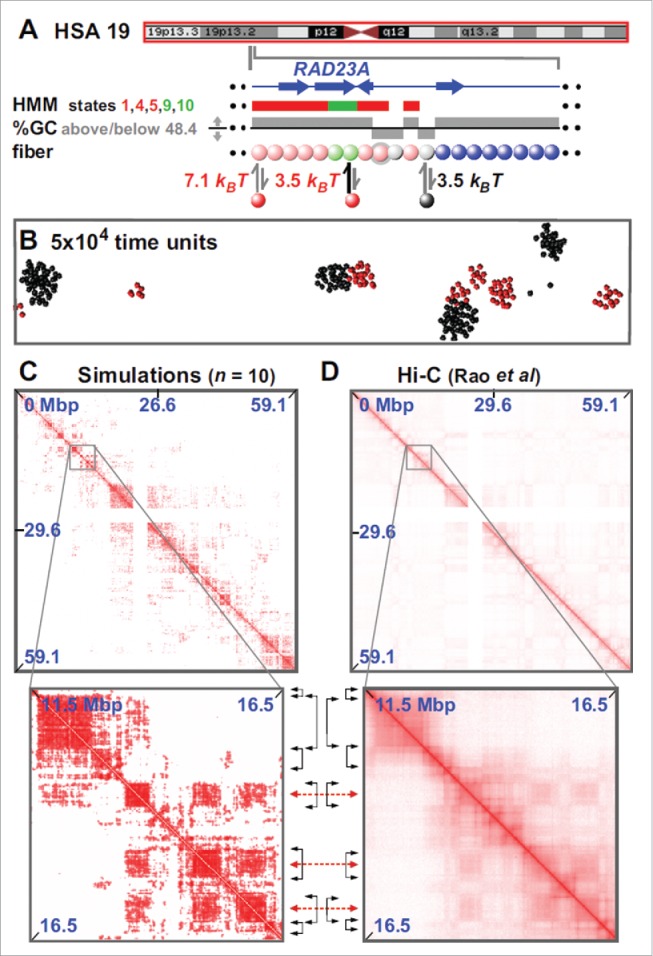
Fitting-free simulations of chromosome 19 in GM12878 cells. (A) Overview. The ideogram (red box indicates the whole chromosome that was simulated) and Broad HMM track (colored regions reflect chromatin states) are from the UCSC browser; the zoom illustrates an arbitrary region, around RAD23A, to show the details of the “coloring.” Beads (3 kbp) are colored according to HMM state and GC content: blue beads are non-binding; pink beads correspond to states 1,4,5 in the ChromHMM track; light-green to states 9,10. Gray beads correspond to beads which have <48.4% GC. Pink and light-green beads bind (respectively, strongly and weakly) active factors (red in the figure); gray beads bind to inactive factors, linked to heterochromatization (black in the figure). Note that the coloring rule is such that beads can have multiple colors: for instance, in the zoom 2 pink beads are also gray (represented by gray halos), so that such beads can bind both red and black factors. (B) Snapshot (without chromatin) of central region after 5×10 4 units; most clusters contain factors (or proteins) of one color. In other words, active and inactive proteins cluster separately. As discussed in the text, the formatio of specialized clusters may underlie both the formation of A/B compartments (when looking at the chromatin interactions) and that of some nuclear bodies (when looking at the protein cluster patterns). (C,D) Comparison between contact maps from simulations and experiments (see Ref.19 for more details). Between zooms, black double-headed arrows mark boundaries of prominent domains (on the diagonal), and red double-headed ones the centers of off-diagonal blocks making many inter-domain contacts. Reproduced from Ref.,19 with permission. © Brackley CA, et al. Reproduced by permission of Brackley CA, et al. Permission to reuse must be obtained from the rightsholder.
