The asymmetric unit comprises one 1,3-bis(3-tert-butyl-2-hydroxy-5-methylbenzyl)-1,3-diazinan-5-ol molecule and one water molecule. The two molecular components are held together through an O—H⋯O hydrogen bond.
Keywords: crystal structure; hexahydropyrimidine; 1,3-diazinane; hydrogen bond
Abstract
In the title hydrate, C28H42N2O3·H2O, the central 1,3-diazinan-5-ol ring adopts a chair conformation with the two benzyl substituents equatorial and the lone pairs of the N atoms axial. The dihedral angle between the aromatic rings is 19.68 (38)°. There are two intramolecular O—H⋯N hydrogen bonds, each generating an S(6) ring motif. In the crystal, classical O—H⋯O hydrogen bonds connect the 1,3-diazinane and water molecules into columns extending along the b axis. The crystal structure was refined as a two-component twin with a fractional contribution to the minor domain of 0.0922 (18).
Chemical context
Current research of our group is directed toward the synthesis of cyclic aminals with conformational interest, which may have the structural requirement for hydrogen-bonded interactions. Obvious targets are the 5-hydroxy-1,3-diazinanes because a hydroxyl group in the six-membered 1,3-diazacyclic ring may alter the conformational preferences resulting from the interactions of the hydroxyl group and the endocyclic nitrogen atoms (Salzner, 1995 ▸). We gradually realized that the structural features of this class of compounds are much more complex than previously believed and defined. Thus, we intend to use X-ray investigations to complement the information on conformational preferences and electronic parameters of 5-hydroxy-1,3-diazinanes obtained using NMR chemical shift data, spin–spin coupling constants, and their NOESY spectra.
We have previously reported the synthesis and crystal structure of 1,3-bis(3-tert-butyl-2-hydroxy-5-methoxybenzyl)-1,3-diazinan-5-ol monohydrate (II) and this study has shown that the hydroxyl substituent on the 1,3-diazinane ring is disordered over two positions, namely one component equatorial and the other axial (Rivera et al., 2014 ▸). As a logical step in the progression of these studies, in this paper we discuss the synthesis and crystal structure of the title compound (I), 1,3-bis(3-tert-butyl-2-hydroxy-5-methylbenzyl)-1,3-diazinan-5-ol monohydrate. The X-ray study again reveals that compound crystallizes with a solvent water molecule that links to the organic molecule through an O—H⋯O hydrogen bond. Furthermore, the hydroxyl group in the pyrimidine ring is also disordered over two positions (axial, equatorial).
Structural commentary
The molecular structure of the title compound is presented in Fig. 1 ▸. The structure consists of a 1,3-bis(3-tert-butyl-2-hydroxy-5-methylbenzyl)-1,3-diazinan-5-ol molecule and a water molecule. These components are connected by an O3—H3⋯O1W hydrogen bond (Table 1 ▸) with the water-O atom as the acceptor. The 1,3-diazinane ring adopts a chair conformation with puckering parameters: Q = 0.588 (2) Å, θ = 176.9 (5) and φ = 245 (9)°. Atoms N1 and N2 are essentially tetrahedral (bond-angle sums are 331.5° for N1 and 331.6° for N2), with their benzyl substituents in equatorial positions and the lone pairs axial. The aromatic rings of these substituents are roughly parallel, with a dihedral angle between the two benzene rings of 19.7 (4)°. Intramolecular O—H⋯N hydrogen bonds form between the pyrimidine N atoms and the OH groups of the benzyl substituents and the pyrimidine N atoms, each with an S(6) graph-set motif (Table 1 ▸). These interactions stabilize the molecular conformation, with O1⋯N1 = 2.696 (5) and O2⋯N2 = 2.702 (5) Å. These distances are closely comparable to those observed in the related structure (II) (Rivera et al., 2014 ▸).
Figure 1.
The molecular structure of the title compound. Displacement ellipsoids are drawn at the 50% probability level. Hydrogen bonds are drawn as dashed lines and, for clarity, only the major-disorder component (equatorial) of the –OH substituent on the pyrimidine ring is included.
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1⋯N1 | 0.95 (7) | 1.84 (7) | 2.696 (5) | 148 (5) |
| O2—H2⋯N2 | 0.96 (6) | 1.81 (6) | 2.702 (5) | 152 (5) |
| O3—H3⋯O1W i | 0.76 (9) | 2.12 (9) | 2.882 (8) | 177 (9) |
| O1W—H1WA⋯O3ii | 0.94 | 1.98 | 2.873 (8) | 158 |
| O1W—H1WA⋯O3′ii | 0.94 | 2.19 | 2.80 (2) | 122 |
| O1W—H1WB⋯O2 | 0.84 | 2.64 | 3.057 (7) | 112 |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
The N2—C7 distance of 1.485 (6) Å is slightly longer than the typical value for an N—C bond [1.469 Å]. The remaining C—N bonds in the molecule are also typical and compare well with those found in the the related structure (II) (Rivera et al., 2014 ▸). The C12—O1 and C22—O2 distances are typical of those for a hydroxy substituent on an aromatic ring [1.376 (6) and 1.374 (5) Å, respectively]. Bond angles within the 1,3-diazinane ring are unexceptional. The hydroxyl group is disordered over two positions, with site occupancies refining to 0.794 (13) and 0.206 (13). The OH group of the major component is in the equatorial position with the minor component axial.
Supramolecular features
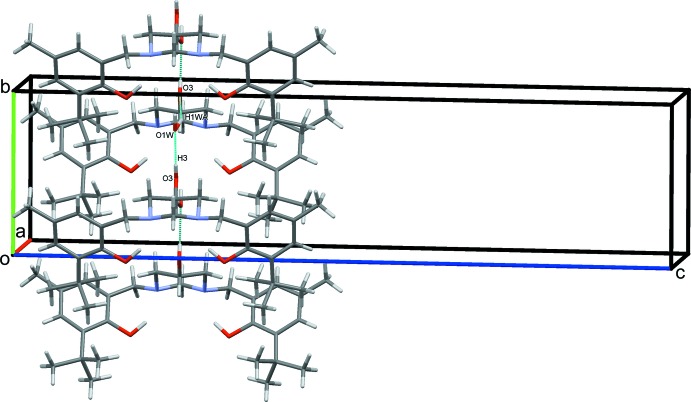
In the crystal, O3—H3⋯O1W hydrogen bonds form chains along b. These contacts are augmented by additional strong O1W—H1WA⋯O3 hydrogen bonds, this time with O3 as the acceptor (Fig. 2 ▸, Table 1 ▸). The chains are held together by van der Waals forces.
Figure 2.
Part of the crystal packing of the title compound, showing the extensive intermolecular hydrogen-bonding interactions (dashed lines). For clarity, only the major-disorder components (equatorial) of the OH substituents on the pyrimidine rings are included.
Database survey
Apart from the previously published structure (Rivera et al., 2014 ▸), there is only one similar entry in the CSD (Mendes et al., 2014 ▸). In this latter structure, the 1,3-diazinane molecule acts as a ligand to an iron(III) cation, which would affect comparisons with the geometric parameters of the title compound.
Synthesis and crystallization
The title compound was prepared according to our reported method (Rivera et al., 2016 ▸). The crude product was recrystallized from hexane solution, giving colorless crystals suitable for X-ray diffraction. M.p. 400 K, yield, 38%.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. The O3—H3 hydroxyl group is disordered over two positions, one with the OH group equatorial with the minor component axial. The site occupancies refine to 0.794 (13) and 0.206 (13), respectively. The H atom of the hydroxyl group of the major component was located in a difference map and refined freely while that of the minor component was fixed geometrically, both with U iso(H) set to 1.2U eq(O). The H atoms of the water molecule were fixed in their found locations with U iso(H) set to 1.5U eq(O). C-bound H atoms were fixed geometrically (C—-H = 0.95 or 0.99 Å) and refined using a riding-model approximation, with U iso(H) set to 1.2U eq of the parent atom. The crystal was a two-component twin with a fractional contribution to the minor domain of 0.0922 (18).
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C28H42N2O3·H2O |
| M r | 472.65 |
| Crystal system, space group | Monoclinic, P21/c |
| Temperature (K) | 173 |
| a, b, c (Å) | 10.11944 (9), 8.25445 (8), 33.8907 (3) |
| β (°) | 97.8676 (4) |
| V (Å3) | 2804.26 (4) |
| Z | 4 |
| Radiation type | Cu Kα |
| μ (mm−1) | 0.59 |
| Crystal size (mm) | 0.25 × 0.25 × 0.09 |
| Data collection | |
| Diffractometer | Bruker APEXII CCD three-circle |
| Absorption correction | Multi-scan (SADABS; Bruker, 1998 ▸) |
| T min, T max | 0.746, 1.000 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 25653, 3138, 2895 |
| R int | 0.053 |
| θmax (°) | 51.7 |
| (sin θ/λ)max (Å−1) | 0.509 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.079, 0.207, 1.07 |
| No. of reflections | 3138 |
| No. of parameters | 333 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.28, −0.31 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989016013645/sj5503sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989016013645/sj5503Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989016013645/sj5503Isup3.cml
CCDC reference: 1500903
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
We acknowledge the Dirección de Investigaciones, Sede Bogotá (DIB) de la Universidad Nacional de Colombia for financial support of this work (research project No. 28427). IMC is also grateful to COLCIENCIAS for his doctoral scholarship.
supplementary crystallographic information
Crystal data
| C28H42N2O3·H2O | F(000) = 1032 |
| Mr = 472.65 | Dx = 1.120 Mg m−3 |
| Monoclinic, P21/c | Cu Kα radiation, λ = 1.54187 Å |
| a = 10.11944 (9) Å | Cell parameters from 9999 reflections |
| b = 8.25445 (8) Å | θ = 2–50° |
| c = 33.8907 (3) Å | µ = 0.59 mm−1 |
| β = 97.8676 (4)° | T = 173 K |
| V = 2804.26 (4) Å3 | Plate, colourless |
| Z = 4 | 0.25 × 0.25 × 0.09 mm |
Data collection
| Bruker APEXII CCD three-circle diffractometer | 2895 reflections with I > 2σ(I) |
| Radiation source: Incoatec microfocus source | Rint = 0.053 |
| ω scans | θmax = 51.7°, θmin = 1.3° |
| Absorption correction: multi-scan (SADABS; Bruker, 1998) | h = −9→10 |
| Tmin = 0.746, Tmax = 1.000 | k = −7→8 |
| 25653 measured reflections | l = −34→34 |
| 3138 independent reflections |
Refinement
| Refinement on F2 | 0 restraints |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.079 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.207 | w = 1/[σ2(Fo2) + (0.0797P)2 + 7.0703P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.07 | (Δ/σ)max < 0.001 |
| 3138 reflections | Δρmax = 0.28 e Å−3 |
| 333 parameters | Δρmin = −0.31 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refined as a 2-component twin. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| N1 | 0.3254 (4) | 0.7763 (5) | 0.21069 (11) | 0.0316 (10) | |
| N2 | 0.3688 (4) | 0.7733 (5) | 0.28166 (11) | 0.0328 (10) | |
| O1 | 0.2304 (4) | 0.5043 (4) | 0.17291 (11) | 0.0455 (10) | |
| H1 | 0.255 (6) | 0.578 (8) | 0.1942 (19) | 0.07 (2)* | |
| O2 | 0.3143 (3) | 0.5008 (4) | 0.32058 (10) | 0.0400 (9) | |
| H2 | 0.329 (5) | 0.575 (7) | 0.2996 (17) | 0.056 (17)* | |
| O3 | 0.0542 (5) | 0.9917 (7) | 0.25116 (16) | 0.0540 (19) | 0.794 (13) |
| H3 | 0.091 (8) | 1.073 (11) | 0.251 (2) | 0.05 (3)* | 0.794 (13) |
| O3' | 0.087 (2) | 0.732 (3) | 0.2516 (8) | 0.084 (10) | 0.206 (13) |
| H3' | 0.1248 | 0.6581 | 0.2401 | 0.126* | 0.206 (13) |
| C1 | 0.4256 (4) | 0.8013 (6) | 0.24530 (13) | 0.0316 (11) | |
| H1A | 0.5012 | 0.7262 | 0.2441 | 0.038* | |
| H1B | 0.4600 | 0.9135 | 0.2451 | 0.038* | |
| C2 | 0.2173 (5) | 0.8935 (6) | 0.21124 (14) | 0.0375 (12) | |
| H2A | 0.1483 | 0.8753 | 0.1880 | 0.045* | |
| H2B | 0.2526 | 1.0047 | 0.2094 | 0.045* | |
| C3 | 0.1561 (5) | 0.8758 (6) | 0.24922 (15) | 0.0379 (13) | |
| H3A | 0.1156 | 0.7654 | 0.2495 | 0.046* | 0.794 (13) |
| H3B | 0.0914 | 0.9667 | 0.2503 | 0.046* | 0.206 (13) |
| C4 | 0.2634 (5) | 0.8907 (6) | 0.28543 (14) | 0.0370 (12) | |
| H4A | 0.3007 | 1.0017 | 0.2870 | 0.044* | |
| H4B | 0.2240 | 0.8695 | 0.3101 | 0.044* | |
| C6 | 0.3872 (5) | 0.7878 (6) | 0.17401 (14) | 0.0378 (13) | |
| H6A | 0.4694 | 0.7213 | 0.1772 | 0.045* | |
| H6B | 0.4135 | 0.9017 | 0.1703 | 0.045* | |
| C7 | 0.4746 (5) | 0.7809 (6) | 0.31659 (14) | 0.0387 (12) | |
| H7A | 0.5061 | 0.8942 | 0.3202 | 0.046* | |
| H7B | 0.5512 | 0.7140 | 0.3110 | 0.046* | |
| C11 | 0.2987 (4) | 0.7334 (5) | 0.13747 (14) | 0.0308 (12) | |
| C12 | 0.2251 (5) | 0.5891 (5) | 0.13778 (14) | 0.0310 (12) | |
| C13 | 0.1507 (5) | 0.5278 (6) | 0.10314 (15) | 0.0377 (13) | |
| C14 | 0.1514 (6) | 0.6207 (7) | 0.06904 (15) | 0.0485 (15) | |
| H14 | 0.1022 | 0.5821 | 0.0450 | 0.058* | |
| C15 | 0.2198 (6) | 0.7672 (7) | 0.06775 (15) | 0.0490 (15) | |
| C16 | 0.2925 (5) | 0.8205 (6) | 0.10255 (15) | 0.0405 (13) | |
| H16 | 0.3398 | 0.9199 | 0.1025 | 0.049* | |
| C17 | 0.0760 (5) | 0.3666 (6) | 0.10303 (17) | 0.0499 (15) | |
| C18 | 0.0022 (8) | 0.3268 (9) | 0.0615 (2) | 0.095 (3) | |
| H18A | −0.0621 | 0.4130 | 0.0530 | 0.143* | |
| H18B | 0.0667 | 0.3184 | 0.0425 | 0.143* | |
| H18C | −0.0452 | 0.2236 | 0.0625 | 0.143* | |
| C19 | 0.1749 (6) | 0.2298 (6) | 0.1152 (2) | 0.071 (2) | |
| H19A | 0.1268 | 0.1267 | 0.1151 | 0.106* | |
| H19B | 0.2399 | 0.2240 | 0.0963 | 0.106* | |
| H19C | 0.2216 | 0.2508 | 0.1420 | 0.106* | |
| C20 | −0.0288 (6) | 0.3723 (7) | 0.1317 (2) | 0.0652 (18) | |
| H20A | −0.0921 | 0.4601 | 0.1239 | 0.098* | |
| H20B | −0.0767 | 0.2689 | 0.1308 | 0.098* | |
| H20C | 0.0156 | 0.3915 | 0.1589 | 0.098* | |
| C21 | 0.4305 (4) | 0.7238 (5) | 0.35448 (14) | 0.0292 (11) | |
| C22 | 0.3553 (4) | 0.5812 (5) | 0.35560 (13) | 0.0259 (11) | |
| C23 | 0.3228 (4) | 0.5197 (6) | 0.39176 (13) | 0.0307 (11) | |
| C24 | 0.3686 (6) | 0.6071 (7) | 0.42536 (15) | 0.0469 (14) | |
| H24 | 0.3490 | 0.5673 | 0.4502 | 0.056* | |
| C25 | 0.4417 (6) | 0.7497 (7) | 0.42547 (16) | 0.0535 (16) | |
| C26 | 0.4698 (5) | 0.8058 (6) | 0.38942 (15) | 0.0431 (14) | |
| H26 | 0.5180 | 0.9043 | 0.3886 | 0.052* | |
| C27 | 0.2443 (5) | 0.3604 (6) | 0.39375 (15) | 0.0370 (13) | |
| C28 | 0.1069 (5) | 0.3741 (7) | 0.36836 (19) | 0.0559 (16) | |
| H28A | 0.0569 | 0.4634 | 0.3783 | 0.084* | |
| H28B | 0.1185 | 0.3950 | 0.3406 | 0.084* | |
| H28C | 0.0577 | 0.2726 | 0.3700 | 0.084* | |
| C29 | 0.3219 (6) | 0.2220 (6) | 0.3783 (2) | 0.0593 (17) | |
| H29A | 0.3377 | 0.2457 | 0.3510 | 0.089* | |
| H29B | 0.4075 | 0.2094 | 0.3954 | 0.089* | |
| H29C | 0.2704 | 0.1215 | 0.3786 | 0.089* | |
| C30 | 0.2210 (8) | 0.3202 (9) | 0.4363 (2) | 0.081 (2) | |
| H30A | 0.1709 | 0.4084 | 0.4467 | 0.121* | |
| H30B | 0.1699 | 0.2193 | 0.4363 | 0.121* | |
| H30C | 0.3071 | 0.3072 | 0.4531 | 0.121* | |
| C151 | 0.2103 (9) | 0.8663 (9) | 0.03030 (18) | 0.090 (2) | |
| H15A | 0.1229 | 0.9194 | 0.0256 | 0.135* | |
| H15B | 0.2211 | 0.7956 | 0.0077 | 0.135* | |
| H15C | 0.2807 | 0.9487 | 0.0332 | 0.135* | |
| C251 | 0.4875 (10) | 0.8412 (10) | 0.4634 (2) | 0.105 (3) | |
| H25A | 0.4431 | 0.9469 | 0.4625 | 0.158* | |
| H25B | 0.5843 | 0.8570 | 0.4660 | 0.158* | |
| H25C | 0.4651 | 0.7793 | 0.4862 | 0.158* | |
| O1W | 0.1841 (5) | 0.3019 (6) | 0.2497 (2) | 0.120 (2) | |
| H1WA | 0.1012 | 0.3526 | 0.2427 | 0.179* | |
| H1WB | 0.2232 | 0.3920 | 0.2488 | 0.179* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N1 | 0.027 (2) | 0.028 (2) | 0.040 (2) | −0.0053 (18) | 0.0046 (18) | −0.0030 (18) |
| N2 | 0.031 (2) | 0.029 (2) | 0.037 (2) | −0.0030 (19) | −0.0001 (18) | 0.0044 (18) |
| O1 | 0.052 (2) | 0.030 (2) | 0.053 (2) | −0.0120 (18) | 0.0012 (18) | 0.0074 (19) |
| O2 | 0.048 (2) | 0.032 (2) | 0.039 (2) | −0.0141 (17) | 0.0033 (16) | −0.0058 (17) |
| O3 | 0.037 (3) | 0.046 (4) | 0.080 (4) | 0.009 (3) | 0.013 (3) | 0.000 (3) |
| O3' | 0.054 (15) | 0.10 (2) | 0.101 (18) | −0.040 (14) | 0.025 (12) | −0.002 (15) |
| C1 | 0.027 (3) | 0.024 (2) | 0.044 (3) | −0.003 (2) | 0.004 (2) | 0.002 (2) |
| C2 | 0.034 (3) | 0.031 (3) | 0.046 (3) | 0.005 (2) | 0.000 (2) | 0.003 (2) |
| C3 | 0.024 (3) | 0.032 (3) | 0.057 (3) | 0.012 (3) | 0.004 (2) | 0.002 (2) |
| C4 | 0.034 (3) | 0.029 (3) | 0.048 (3) | 0.004 (2) | 0.009 (2) | 0.002 (2) |
| C6 | 0.036 (3) | 0.033 (3) | 0.046 (3) | −0.011 (2) | 0.010 (2) | 0.000 (2) |
| C7 | 0.034 (3) | 0.030 (3) | 0.050 (3) | −0.006 (2) | −0.005 (2) | 0.003 (2) |
| C11 | 0.030 (3) | 0.019 (3) | 0.044 (3) | −0.004 (2) | 0.010 (2) | −0.006 (2) |
| C12 | 0.028 (3) | 0.020 (3) | 0.045 (3) | 0.004 (2) | 0.007 (2) | 0.003 (2) |
| C13 | 0.035 (3) | 0.026 (3) | 0.052 (3) | 0.002 (2) | 0.005 (2) | −0.010 (2) |
| C14 | 0.052 (4) | 0.051 (4) | 0.040 (3) | 0.001 (3) | −0.001 (3) | −0.014 (3) |
| C15 | 0.065 (4) | 0.044 (4) | 0.040 (3) | −0.002 (3) | 0.014 (3) | −0.001 (3) |
| C16 | 0.051 (3) | 0.030 (3) | 0.043 (3) | −0.004 (3) | 0.016 (3) | 0.001 (2) |
| C17 | 0.038 (3) | 0.036 (3) | 0.073 (4) | −0.004 (3) | −0.001 (3) | −0.016 (3) |
| C18 | 0.106 (6) | 0.072 (5) | 0.100 (6) | −0.043 (5) | −0.015 (5) | −0.030 (4) |
| C19 | 0.046 (4) | 0.021 (3) | 0.148 (6) | −0.001 (3) | 0.022 (4) | −0.017 (3) |
| C20 | 0.035 (3) | 0.041 (4) | 0.121 (5) | −0.004 (3) | 0.016 (4) | −0.006 (3) |
| C21 | 0.022 (3) | 0.018 (3) | 0.044 (3) | −0.002 (2) | −0.007 (2) | −0.004 (2) |
| C22 | 0.023 (3) | 0.017 (3) | 0.035 (3) | 0.004 (2) | −0.007 (2) | −0.008 (2) |
| C23 | 0.026 (3) | 0.028 (3) | 0.038 (3) | 0.005 (2) | 0.003 (2) | −0.002 (2) |
| C24 | 0.059 (4) | 0.046 (3) | 0.036 (3) | −0.003 (3) | 0.009 (3) | −0.005 (3) |
| C25 | 0.068 (4) | 0.045 (4) | 0.044 (3) | −0.008 (3) | −0.002 (3) | −0.021 (3) |
| C26 | 0.046 (3) | 0.027 (3) | 0.053 (4) | −0.007 (3) | −0.006 (3) | −0.010 (3) |
| C27 | 0.027 (3) | 0.030 (3) | 0.055 (3) | 0.001 (2) | 0.010 (2) | 0.005 (2) |
| C28 | 0.031 (3) | 0.040 (3) | 0.098 (5) | −0.004 (3) | 0.014 (3) | 0.003 (3) |
| C29 | 0.048 (4) | 0.023 (3) | 0.110 (5) | −0.003 (3) | 0.022 (3) | 0.002 (3) |
| C30 | 0.103 (6) | 0.069 (5) | 0.073 (4) | −0.024 (4) | 0.023 (4) | 0.014 (4) |
| C151 | 0.132 (7) | 0.088 (5) | 0.048 (4) | −0.009 (5) | 0.010 (4) | 0.016 (4) |
| C251 | 0.157 (8) | 0.097 (6) | 0.059 (4) | −0.050 (6) | 0.006 (5) | −0.036 (4) |
| O1W | 0.081 (4) | 0.058 (3) | 0.220 (7) | −0.002 (3) | 0.022 (4) | −0.010 (4) |
Geometric parameters (Å, º)
| N1—C1 | 1.456 (6) | C17—C20 | 1.535 (8) |
| N1—C2 | 1.462 (6) | C17—C18 | 1.536 (8) |
| N1—C6 | 1.469 (6) | C18—H18A | 0.9800 |
| N2—C1 | 1.448 (6) | C18—H18B | 0.9800 |
| N2—C4 | 1.460 (6) | C18—H18C | 0.9800 |
| N2—C7 | 1.485 (6) | C19—H19A | 0.9800 |
| O1—C12 | 1.376 (6) | C19—H19B | 0.9800 |
| O1—H1 | 0.95 (7) | C19—H19C | 0.9800 |
| O2—C22 | 1.374 (5) | C20—H20A | 0.9800 |
| O2—H2 | 0.96 (6) | C20—H20B | 0.9800 |
| O3—C3 | 1.415 (6) | C20—H20C | 0.9800 |
| O3—H3 | 0.76 (9) | C21—C26 | 1.375 (7) |
| O3'—C3 | 1.38 (2) | C21—C22 | 1.405 (6) |
| O3'—H3' | 0.8400 | C22—C23 | 1.407 (6) |
| C1—H1A | 0.9900 | C23—C24 | 1.374 (7) |
| C1—H1B | 0.9900 | C23—C27 | 1.542 (7) |
| C2—C3 | 1.510 (7) | C24—C25 | 1.390 (8) |
| C2—H2A | 0.9900 | C24—H24 | 0.9500 |
| C2—H2B | 0.9900 | C25—C26 | 1.372 (8) |
| C3—C4 | 1.528 (7) | C25—C251 | 1.507 (8) |
| C3—H3A | 1.0000 | C26—H26 | 0.9500 |
| C3—H3B | 1.0000 | C27—C29 | 1.519 (7) |
| C4—H4A | 0.9900 | C27—C30 | 1.529 (8) |
| C4—H4B | 0.9900 | C27—C28 | 1.536 (7) |
| C6—C11 | 1.494 (7) | C28—H28A | 0.9800 |
| C6—H6A | 0.9900 | C28—H28B | 0.9800 |
| C6—H6B | 0.9900 | C28—H28C | 0.9800 |
| C7—C21 | 1.493 (7) | C29—H29A | 0.9800 |
| C7—H7A | 0.9900 | C29—H29B | 0.9800 |
| C7—H7B | 0.9900 | C29—H29C | 0.9800 |
| C11—C16 | 1.378 (7) | C30—H30A | 0.9800 |
| C11—C12 | 1.406 (6) | C30—H30B | 0.9800 |
| C12—C13 | 1.400 (7) | C30—H30C | 0.9800 |
| C13—C14 | 1.388 (7) | C151—H15A | 0.9800 |
| C13—C17 | 1.530 (7) | C151—H15B | 0.9800 |
| C14—C15 | 1.397 (8) | C151—H15C | 0.9800 |
| C14—H14 | 0.9500 | C251—H25A | 0.9800 |
| C15—C16 | 1.374 (7) | C251—H25B | 0.9800 |
| C15—C151 | 1.502 (8) | C251—H25C | 0.9800 |
| C16—H16 | 0.9500 | O1W—H1WA | 0.9381 |
| C17—C19 | 1.528 (8) | O1W—H1WB | 0.8447 |
| C1—N1—C2 | 109.6 (4) | C20—C17—C18 | 107.2 (5) |
| C1—N1—C6 | 110.0 (3) | C17—C18—H18A | 109.5 |
| C2—N1—C6 | 111.9 (4) | C17—C18—H18B | 109.5 |
| C1—N2—C4 | 110.4 (4) | H18A—C18—H18B | 109.5 |
| C1—N2—C7 | 110.2 (4) | C17—C18—H18C | 109.5 |
| C4—N2—C7 | 111.0 (4) | H18A—C18—H18C | 109.5 |
| C12—O1—H1 | 108 (4) | H18B—C18—H18C | 109.5 |
| C22—O2—H2 | 106 (3) | C17—C19—H19A | 109.5 |
| C3—O3—H3 | 104 (6) | C17—C19—H19B | 109.5 |
| C3—O3'—H3' | 109.5 | H19A—C19—H19B | 109.5 |
| N2—C1—N1 | 110.5 (3) | C17—C19—H19C | 109.5 |
| N2—C1—H1A | 109.6 | H19A—C19—H19C | 109.5 |
| N1—C1—H1A | 109.6 | H19B—C19—H19C | 109.5 |
| N2—C1—H1B | 109.6 | C17—C20—H20A | 109.5 |
| N1—C1—H1B | 109.6 | C17—C20—H20B | 109.5 |
| H1A—C1—H1B | 108.1 | H20A—C20—H20B | 109.5 |
| N1—C2—C3 | 110.0 (4) | C17—C20—H20C | 109.5 |
| N1—C2—H2A | 109.7 | H20A—C20—H20C | 109.5 |
| C3—C2—H2A | 109.7 | H20B—C20—H20C | 109.5 |
| N1—C2—H2B | 109.7 | C26—C21—C22 | 118.9 (4) |
| C3—C2—H2B | 109.7 | C26—C21—C7 | 120.0 (4) |
| H2A—C2—H2B | 108.2 | C22—C21—C7 | 121.0 (4) |
| O3'—C3—C2 | 113.6 (11) | O2—C22—C21 | 118.8 (4) |
| O3—C3—C2 | 111.2 (4) | O2—C22—C23 | 119.9 (4) |
| O3'—C3—C4 | 109.2 (12) | C21—C22—C23 | 121.2 (4) |
| O3—C3—C4 | 110.5 (4) | C24—C23—C22 | 116.1 (4) |
| C2—C3—C4 | 110.4 (4) | C24—C23—C27 | 121.9 (4) |
| O3—C3—H3A | 108.2 | C22—C23—C27 | 122.0 (4) |
| C2—C3—H3A | 108.2 | C23—C24—C25 | 124.5 (5) |
| C4—C3—H3A | 108.2 | C23—C24—H24 | 117.8 |
| O3'—C3—H3B | 107.8 | C25—C24—H24 | 117.8 |
| C2—C3—H3B | 107.8 | C26—C25—C24 | 117.3 (5) |
| C4—C3—H3B | 107.8 | C26—C25—C251 | 120.8 (6) |
| N2—C4—C3 | 108.9 (4) | C24—C25—C251 | 121.9 (6) |
| N2—C4—H4A | 109.9 | C25—C26—C21 | 122.0 (5) |
| C3—C4—H4A | 109.9 | C25—C26—H26 | 119.0 |
| N2—C4—H4B | 109.9 | C21—C26—H26 | 119.0 |
| C3—C4—H4B | 109.9 | C29—C27—C30 | 108.3 (5) |
| H4A—C4—H4B | 108.3 | C29—C27—C28 | 109.5 (5) |
| N1—C6—C11 | 114.0 (4) | C30—C27—C28 | 107.4 (5) |
| N1—C6—H6A | 108.8 | C29—C27—C23 | 109.4 (4) |
| C11—C6—H6A | 108.8 | C30—C27—C23 | 111.9 (4) |
| N1—C6—H6B | 108.8 | C28—C27—C23 | 110.2 (4) |
| C11—C6—H6B | 108.8 | C27—C28—H28A | 109.5 |
| H6A—C6—H6B | 107.7 | C27—C28—H28B | 109.5 |
| N2—C7—C21 | 113.8 (4) | H28A—C28—H28B | 109.5 |
| N2—C7—H7A | 108.8 | C27—C28—H28C | 109.5 |
| C21—C7—H7A | 108.8 | H28A—C28—H28C | 109.5 |
| N2—C7—H7B | 108.8 | H28B—C28—H28C | 109.5 |
| C21—C7—H7B | 108.8 | C27—C29—H29A | 109.5 |
| H7A—C7—H7B | 107.7 | C27—C29—H29B | 109.5 |
| C16—C11—C12 | 119.1 (4) | H29A—C29—H29B | 109.5 |
| C16—C11—C6 | 120.4 (4) | C27—C29—H29C | 109.5 |
| C12—C11—C6 | 120.4 (4) | H29A—C29—H29C | 109.5 |
| O1—C12—C13 | 119.6 (4) | H29B—C29—H29C | 109.5 |
| O1—C12—C11 | 118.7 (4) | C27—C30—H30A | 109.5 |
| C13—C12—C11 | 121.6 (4) | C27—C30—H30B | 109.5 |
| C14—C13—C12 | 115.8 (4) | H30A—C30—H30B | 109.5 |
| C14—C13—C17 | 122.5 (5) | C27—C30—H30C | 109.5 |
| C12—C13—C17 | 121.7 (5) | H30A—C30—H30C | 109.5 |
| C13—C14—C15 | 124.3 (5) | H30B—C30—H30C | 109.5 |
| C13—C14—H14 | 117.8 | C15—C151—H15A | 109.5 |
| C15—C14—H14 | 117.8 | C15—C151—H15B | 109.5 |
| C16—C15—C14 | 117.3 (5) | H15A—C151—H15B | 109.5 |
| C16—C15—C151 | 121.0 (5) | C15—C151—H15C | 109.5 |
| C14—C15—C151 | 121.6 (5) | H15A—C151—H15C | 109.5 |
| C15—C16—C11 | 121.8 (5) | H15B—C151—H15C | 109.5 |
| C15—C16—H16 | 119.1 | C25—C251—H25A | 109.5 |
| C11—C16—H16 | 119.1 | C25—C251—H25B | 109.5 |
| C19—C17—C13 | 109.7 (4) | H25A—C251—H25B | 109.5 |
| C19—C17—C20 | 109.6 (5) | C25—C251—H25C | 109.5 |
| C13—C17—C20 | 110.8 (4) | H25A—C251—H25C | 109.5 |
| C19—C17—C18 | 107.9 (5) | H25B—C251—H25C | 109.5 |
| C13—C17—C18 | 111.5 (5) | H1WA—O1W—H1WB | 90.3 |
| C4—N2—C1—N1 | −63.3 (5) | C151—C15—C16—C11 | 177.9 (6) |
| C7—N2—C1—N1 | 173.7 (4) | C12—C11—C16—C15 | −2.2 (7) |
| C2—N1—C1—N2 | 62.5 (5) | C6—C11—C16—C15 | 175.2 (5) |
| C6—N1—C1—N2 | −174.0 (4) | C14—C13—C17—C19 | 118.1 (6) |
| C1—N1—C2—C3 | −58.2 (5) | C12—C13—C17—C19 | −60.6 (6) |
| C6—N1—C2—C3 | 179.4 (4) | C14—C13—C17—C20 | −120.7 (6) |
| N1—C2—C3—O3' | −68.0 (13) | C12—C13—C17—C20 | 60.6 (6) |
| N1—C2—C3—O3 | 178.0 (4) | C14—C13—C17—C18 | −1.4 (7) |
| N1—C2—C3—C4 | 55.0 (5) | C12—C13—C17—C18 | 179.9 (5) |
| C1—N2—C4—C3 | 58.5 (5) | N2—C7—C21—C26 | −139.0 (4) |
| C7—N2—C4—C3 | −179.0 (4) | N2—C7—C21—C22 | 44.8 (6) |
| O3'—C3—C4—N2 | 70.9 (11) | C26—C21—C22—O2 | 179.0 (4) |
| O3—C3—C4—N2 | −178.0 (4) | C7—C21—C22—O2 | −4.8 (6) |
| C2—C3—C4—N2 | −54.7 (5) | C26—C21—C22—C23 | −1.5 (6) |
| C1—N1—C6—C11 | 169.1 (4) | C7—C21—C22—C23 | 174.7 (4) |
| C2—N1—C6—C11 | −68.8 (5) | O2—C22—C23—C24 | 179.6 (4) |
| C1—N2—C7—C21 | −169.3 (4) | C21—C22—C23—C24 | 0.1 (6) |
| C4—N2—C7—C21 | 68.0 (5) | O2—C22—C23—C27 | 1.5 (6) |
| N1—C6—C11—C16 | 137.4 (5) | C21—C22—C23—C27 | −178.0 (4) |
| N1—C6—C11—C12 | −45.3 (6) | C22—C23—C24—C25 | 0.8 (8) |
| C16—C11—C12—O1 | −179.0 (4) | C27—C23—C24—C25 | 178.9 (5) |
| C6—C11—C12—O1 | 3.6 (6) | C23—C24—C25—C26 | −0.2 (9) |
| C16—C11—C12—C13 | 3.0 (7) | C23—C24—C25—C251 | 179.2 (6) |
| C6—C11—C12—C13 | −174.3 (4) | C24—C25—C26—C21 | −1.3 (8) |
| O1—C12—C13—C14 | −179.7 (4) | C251—C25—C26—C21 | 179.3 (6) |
| C11—C12—C13—C14 | −1.8 (7) | C22—C21—C26—C25 | 2.2 (7) |
| O1—C12—C13—C17 | −0.9 (7) | C7—C21—C26—C25 | −174.1 (5) |
| C11—C12—C13—C17 | 177.0 (4) | C24—C23—C27—C29 | −117.7 (5) |
| C12—C13—C14—C15 | −0.3 (8) | C22—C23—C27—C29 | 60.2 (6) |
| C17—C13—C14—C15 | −179.1 (5) | C24—C23—C27—C30 | 2.3 (7) |
| C13—C14—C15—C16 | 1.1 (8) | C22—C23—C27—C30 | −179.7 (5) |
| C13—C14—C15—C151 | −176.6 (6) | C24—C23—C27—C28 | 121.8 (5) |
| C14—C15—C16—C11 | 0.2 (8) | C22—C23—C27—C28 | −60.2 (6) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1···N1 | 0.95 (7) | 1.84 (7) | 2.696 (5) | 148 (5) |
| O2—H2···N2 | 0.96 (6) | 1.81 (6) | 2.702 (5) | 152 (5) |
| O3—H3···O1Wi | 0.76 (9) | 2.12 (9) | 2.882 (8) | 177 (9) |
| O1W—H1WA···O3ii | 0.94 | 1.98 | 2.873 (8) | 158 |
| O1W—H1WA···O3′ii | 0.94 | 2.19 | 2.80 (2) | 122 |
| O1W—H1WB···O2 | 0.84 | 2.64 | 3.057 (7) | 112 |
Symmetry codes: (i) x, y+1, z; (ii) −x, y−1/2, −z+1/2.
References
- Bruker (1998). SAINT and SADABS. Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruker (2004). APEX2. Bruker AXS Inc., Madison, Wisconsin, USA.
- Mendes, L. L., Fernandes, C., Franco, R. W. A., Lube, L. M., Wei, S.-H., Reibenspies, J. H., Darnesbourg, D. J. & Horn, A. Jr (2014). J. Braz. Chem. Soc. 25, 1050–1061.
- Rivera, A., Miranda-Carvajal, I., Osorio, H. J., Ríos-Motta, J. & Bolte, M. (2014). Acta Cryst. E70, o687–o688. [DOI] [PMC free article] [PubMed]
- Rivera, A., Miranda-Carvajal, I. & Ríos-Motta, J. (2016). J. Chil. Chem. Soc. Accepted (Paper number, 4317).
- Salzner, U. (1995). J. Org. Chem. 60, 986–995.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989016013645/sj5503sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989016013645/sj5503Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989016013645/sj5503Isup3.cml
CCDC reference: 1500903
Additional supporting information: crystallographic information; 3D view; checkCIF report