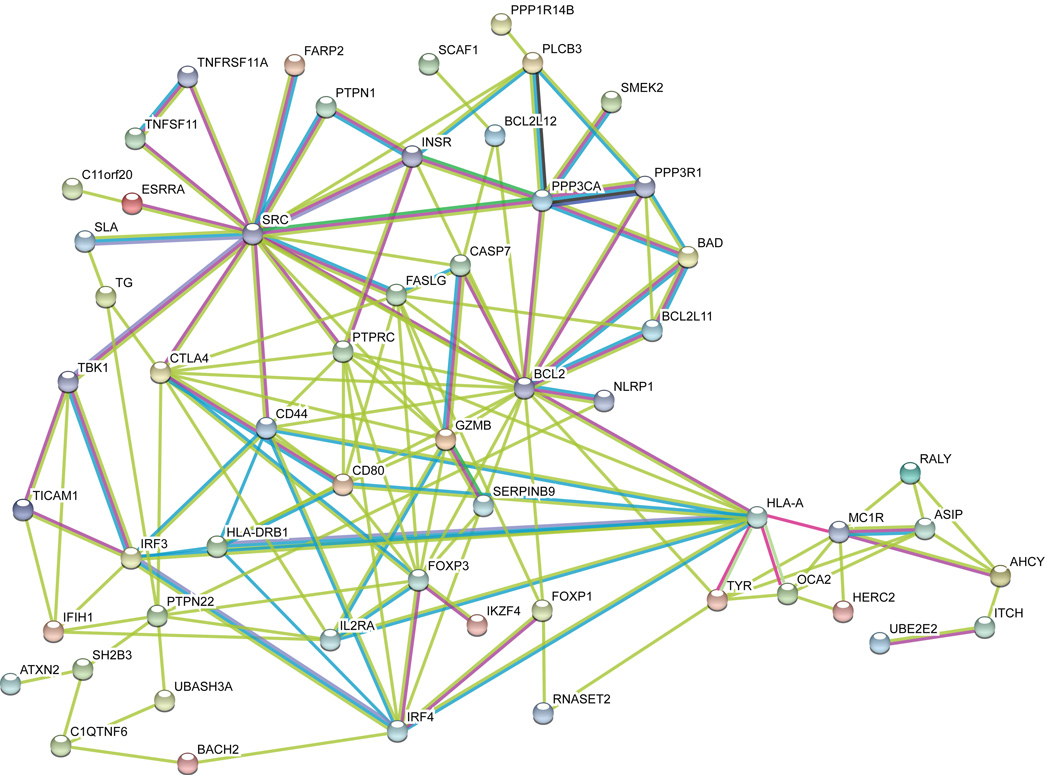
Figure 2.
Bioinformatic functional interaction network analysis of proteins encoded by all positional candidate genes at all confirmed and suggestive vitiligo candidate loci. As a first step, unsupervised functional interaction network analysis was carried out using STRING v10.011, considering each protein as a node and permitting ≤ 5 second-order interactions to maximize connectivity. Nodes that shared no edges with other nodes were then excluded from the network. Edge colors are per STRING: teal, interactions from curated databases; purple, experimentally determined interactions; green, gene neighborhood; blue, databases; red, gene fusions; dark blue, gene co-occurrence; pale green, text-mining; black, co-expression; lavender, protein homology. Note that SMEK2 is an alternative name for PPP4R3B.