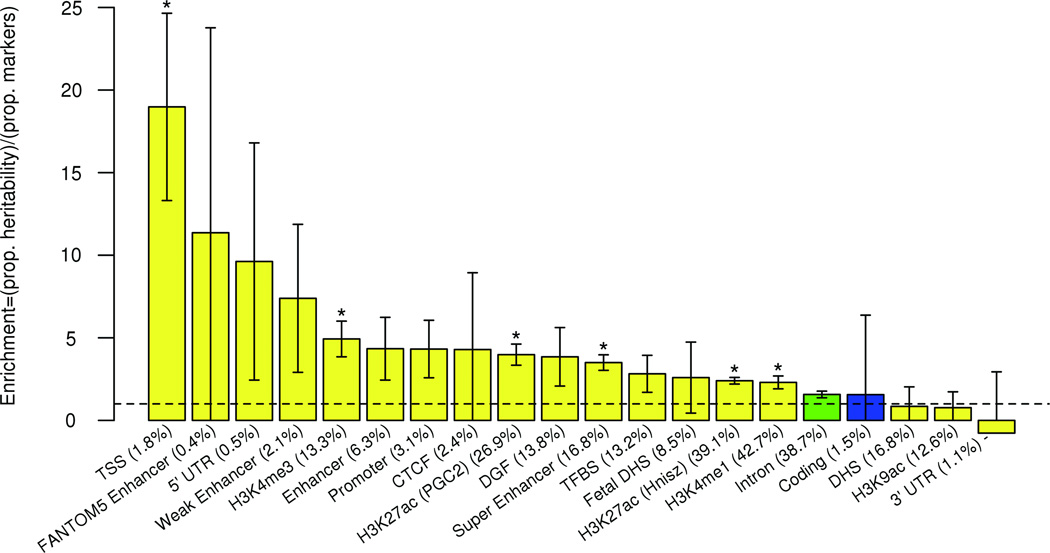
Figure 4.
Enrichment estimates for functional annotations. The combined CMH GWAS123 summary statistics were analyzed using the stratified LD score regression method utilizing the full baseline model51. Regulatory, yellow; protein coding, blue; intron, green. Bar height represents enrichment which is defined to be the proportion of SNP heritability in the category divided by the proportion of SNPs in that category. Error bars represent jackknife standard error around the enrichment. For each category, percentage of the total markers in the category is in parentheses. Dashed line represents a ratio of 1 (no enrichment). Asterisks indicate enrichment significant at P < 0.05 after Bonferroni correction for the 20 categories tested (the categories conserved, repressed, transcribed, and promoter flanking were removed and considered insufficiently specific). CTCF, CCCTC-binding factor; DGF, digital genomic footprint; DHS, DNase hypersensitivity site; TFBS, transcription factor binding site; TSS, transcriptional start site; 5’ and 3’ UTR, 5’ and 3’ untranslated regions. H3K4me1, H3K4me3, H3K9ac, and H3K27ac are regulatory chromatin marks56,57.