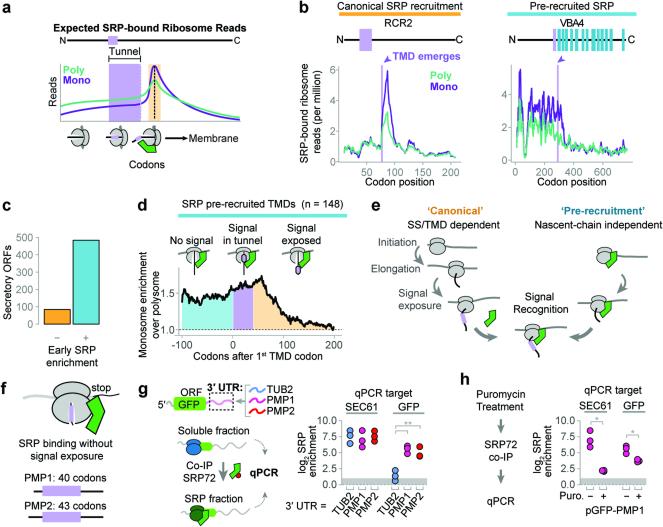
Figure 3. Distinct mechanisms of SRP recruitment.
a, Recruitment of SRP to RNCs is expected to increase ribosome-protected reads from SRP-bound monosomes when an SS or TMD is exposed to the cytosol (orange). b, Distributions of SRP-bound ribosome reads on representative transcripts from cycloheximide-treated cultures. Selected transcripts are RCR2, and VBA4. c, The majority of secretory proteins demonstrated SRP enrichment prior to signal exposure. d, Metagene plot of the median value of enrichment of SRP-bound monosomes over polysomes. Included transcripts encode TMDs at least 40 codons from the start codon. Shaded areas represent enrichment before the TMD is encoded (cyan), while the TMD is in the ribosome exit tunnel (lavender), and after the TMD is exposed (orange). e, Two mechanisms for SRP to select secretory mRNA. f, PMP1 and PMP2 were the only TA proteins that enriched SRP. g, The GFP ORF was fused to the indicated 3′ UTRs and expressed in vivo. Srp72p-TAP was immunoprecipitated from the total soluble fraction and RNAs were subject to qPCR. **p ≤ 0.01, n = 3 biological replicates, Welch's t-test. h, Puromycin treatment of lysate from yeast expressing GFP with the PMP1 3′ UTR was followed by SRP immunoprecipitation and qPCR. *p ≤ 0.05, n = 3 biological replicates, Welch's t-test.