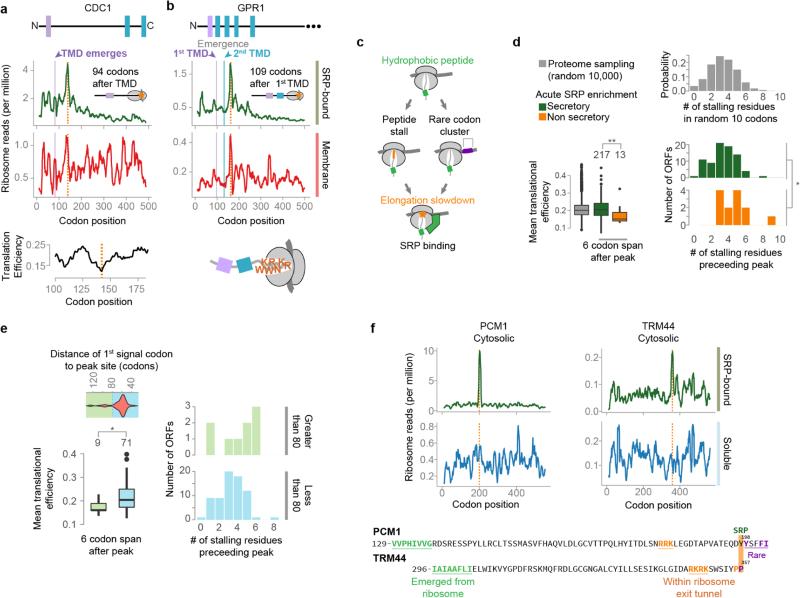
Extended Data Figure 3. Elongation pausing and local SRP recruitment.
a, b, Local increases of ribosome-protected reads from membrane-bound polysomes, indicated with orange lines, were coincident with rare codons, as in cell division cycle protein 1 (CDC1, a) or polybasic nascent chains, as in the plasma membrane G-protein coupled receptor (GPR1, b). Soluble SRP-bound polysome-protected reads were further increased at the same positions. c, In these cases, hydrophobic sequences in the nascent chain were exposed to the cytosol at the locations of increased reads, which were coincident with elongation attenuators. d, Translational efficiencies for the 6 codons following, and the number of stalling residues within the 10 residues preceding, the sites of increased SRP-bound ribosome reads. Translational efficiency was determined by attributing the nTE score to each codon55. Residues that were found to stall the ribosome, based on previous investigation20,56,57, were lysine, arginine, glutamate, aspartate, proline and glycine. Because of variation in specific motifs, and uncertainty in whether these motifs are additive, we simply compared the total number of these residues in the indicated 10 residue spans. Sets of 10,000 random sequences, at least 10 amino acids from the stop codon, were sampled from 5907 non-dubious ORFs, and translational efficiency and stalling residues were determined over 6 or 10 codon spans. *p ≤ 0.05, **p ≤ 0.01, Wilcoxon rank-sum tests. e, The targeting signals that recruited SRP directly to the nascent chain unusually far from the encoding of the signal had SRP-binding sites coincident with intrinsic elongation attenuation. Secretory protein transcripts that showed an increase in SRP-bound protected reads (see Extended Data Fig. 2c, f) were further classified by the position of the peak relative to the first signal codon. Transcripts with peaks found at least 80 codons after the signal had significantly lower translational efficiency in the 6 codons following the peak. These transcripts also had a greater, but not statistically significant, amount of stalling amino acids in the 10 residues preceding the peak. *p ≤ 0.05, Wilcoxon rank-sum tests. f, Similar increases in SRP-bound reads were observed for certain non-secretory proteins as exemplified by phosphoacetylglucosamine mutase (PCM1) and tRNASer Um44 2′-O-methyltransferase (TRM44). Hydrophobic sequences in non-secretory proteins, coupled with attenuation of elongation, may lead to SRP recruitment.