INTRODUCTION
With advancements in pharmacogenomics research and genotyping technology, implementation of pharmacogenomics into clinical practice is now feasible. The aim of this publication is to serve as a tutorial for institutions interested in developing pharmacogenomics services. Topics covered include resources needed, clinical decision support establishment, choosing a genotyping platform, and challenges faced with pharmacogenomics service implementation. This tutorial provides practical advice, drawing upon experience of two established clinical pharmacogenomics services.
BACKGROUND
Clinical characteristics, such as age, body weight, and renal and liver function, have long been used by clinicians to inform drug selection and dosing in clinical practice. Despite this practice, variability in medication response (including lack of efficacy and adverse events) is still commonly observed in patients. The effect of an individual's genomic profile on his/her drug response, or pharmacogenomics (PGx), is thought to predict between 20% and 95% of response variability, depending on the drug.1, 2, 3, 4 Thus, combining genetic information with clinical data is an intervention that may improve the safety and effectiveness of medication therapy. Although the benefit of using PGx data to optimize medication therapy is recognized by major stakeholders in the healthcare system, including patients, healthcare professionals, and healthcare insurers,5 its integration into routine clinical practice has been relatively slow compared with the pace of advancement in PGx knowledge and genotyping technology.
A number of medical centers in the United States have implemented clinical PGx services in recent years.6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18 The genotyping methods used by these services can be categorized into two groups: (i) preemptive clinical genotyping; and (ii) reactive clinical genotyping. Preemptive clinical genotyping likely represents the most efficient method of PGx data utilization, as genotypes for multiple prespecified variations related to several medications are measured simultaneously. Genetic information is stored in the electronic health record (EHR) and can be used when patients are started on drugs where this information would be helpful. Because of the economies of scale used in preemptive genotyping, the overall cost to genotype variants in many genes simultaneously may be lower than the cost to test variants in each gene individually. On the other hand, reactive genotyping tests focus on the specific variation(s) related to the drug being started at that time. A single test can often be run sooner than a larger preemptive genotyping panel, which are often run in batches to remain cost‐competitive. Because of the issues faced with incorporating numerous PGx results (currently actionable vs. potentially actionable in the future) into the EHR, the reduced likelihood of insurance reimbursement, and the large startup costs associated with preemptive genotyping, reactive genotyping might be a more feasible method to use when starting clinical PGx implementation, especially in institutions with limited personnel and financial support.
In President Obama's State of the Union address in January 2015, he announced the Precision Medicine Initiative, which will focus on prevention and treatment approaches that account for interindividual variability in drug response, which largely includes PGx. Although the majority of institutions that are currently implementing clinical PGx services in the United States are large academic medical centers, the Precision Medicine Initiative may signal it is time for other institutions to consider implementing as well. Thus, we have created a tutorial for interested clinicians that discuss various approaches to implementing a reactive clinical PGx service, including selection of gene‐drug pairs, administrative considerations, and data collection, along with insight and practical advice from two established clinical PGx services at the University of Illinois at Chicago (UIC) and University of Florida (UF) medical centers. Basic PGx concepts have previously been reviewed,19, 20, 21, 22 and may provide a useful background for those needing it. Other resources available to enhance education on genomic competencies include Global Genetics and Genomics Community (http://g‐3‐c.org/en) and the National Institutes of Health (NIH) Genetics/Genomics Competency Center (http://g‐2‐c‐2.org//).
SELECTING GENE‐DRUG PAIRS FOR IMPLEMENTATION
Before assessing the capabilities and preparedness of the institution's facilities and personnel, a first gene‐drug pair must be identified for implementation. Ideally, the gene(s) being tested would provide information relating to the drug's safety or efficacy, and should be chosen with a target group of patients in mind. At this point, a physician champion is key in advocating for specific gene‐drug pairs to implement. Early selection of the initial gene‐drug pair(s) will aid in the development of a framework for PGx service implementation. Several factors must be considered when selecting which gene‐drug pair(s) to implement. First, the PGx test must have enough evidence to support its clinical utility. Gene‐drug pairs with evidence‐based guidelines, such as those issued by the Clinical Pharmacogenetics Implementation Consortium (CPIC), and with clear clinical recommendations make good candidates. Second, it is important to implement a PGx test that is relevant to the institution's patient population with regard to frequency of variation within the selected gene(s). Although common genetic variation might seem to be an obvious choice, less common variants that are associated with more severe outcomes (particularly with regard to adverse effects or untreated disease) are also meaningful gene‐drug pairs to implement. For instance, providing HLA‐B genetic testing for carbamazepine use at an institution that rarely treats anyone of Southeastern Asian descent (almost exclusively in whom the variant associated with hypersensitivity is observed) would not likely be cost‐effective nor benefit the institution's patient population as a whole. Additionally, information on the frequency of selected genetic variants will help in estimating the potential service workload. Table 1 displays variant allele frequencies of PGx implementation candidates among different ethnic/racial groups. Last, other considerations include the ability of instruments/testing platform to genotype all variants of interest within the gene(s), the presence of personnel with sufficient expertise with the drug and disease state involved, and the likelihood of insurance reimbursement for a specific test.
Table 1.
| Allele frequency | |||||
|---|---|---|---|---|---|
| Drug | Allele | African | American | Asian | European |
| Abacavir | HLA‐B*57:01 | 1% | 2.6% (South American) | 1.6% | 6.8% |
| Carbamazepine | HLA‐B*15:02 | 0% | 0.39% (American, non‐white) | 4.3% (East Asian) | <0.1% |
| Clopidogrel |
|
|
|
|
|
| Codeine |
|
|
|
|
|
| Tacrolimus |
|
|
|
|
|
| Thiopurines |
|
|
|
|
|
| Warfarin |
|
|
|
|
|
Six gene‐drug pairs that institutions may want to consider to implement first in their reactive PGx service are: HLA‐B and abacavir and carbamazepine; CYP2C19 and clopidogrel; TPMT and azathioprine, mercaptopurine, and thioguanine; CYP3A5 and tacrolimus; CYP2D6 and opioids; and CYP2C9/VKORC1 and warfarin. Many of these pairs (Table 2) are already being implemented at large academic institutions, with guidelines for their use freely accessible through the Pharmacogenomics Research Network and Pharmacogenomics Knowledge Base website (https://cpicpgx.org/ or https://www.pharmgkb.org/page/cpic). A majority of the aforementioned genes (other than HLA‐B and VKORC1) encode drug metabolizing enzymes, and each individual's genotype for a particular gene can be categorized into one of the five phenotypes that describe the enzyme's activity: ultrarapid metabolizer, rapid metabolizer, extensive (normal) metabolizer, intermediate metabolizer, and poor metabolizer (Table 3). As with utilizing any laboratory test result to make clinical decisions, the clinician cannot solely rely on the results of the test, but must also consider patient‐specific factors, including those that affect the drug's pharmacokinetics (including renal and hepatic dysfunction and drug‐drug interactions) when making specific clinical recommendations based on the patient's genomic information.
Table 2.
| Gene | Drug | |
|---|---|---|
| CFTR | Ivacaftor | |
| CYP2C19 |
|
|
| CYP2C9/VKORC1 | Warfarin | |
| CYP2D6 |
|
|
| CYP3A5 | Tacrolimus | |
| DYPD |
|
|
| G6PD | Rasburicase | |
| HLA‐B*15:02 |
|
|
| HLA‐B*57:01 | Abacavir | |
| HLA‐B*58:01 | Allopurinol | |
| IFNL3 (IL28B) |
|
|
| SLCO1B1 | Simvastatin | |
| TPMT |
|
|
Drug label does not contain recommendation for genotyping, but alteration in efficacy, dosage, or toxicity of the drug by genetic variation is mentioned.
Only gene‐drug pairs with available Clinical Pharmacogenetics Implementation Consortium guidelines are included.
Table 3.
General phenotype definitions for drug‐metabolizing enzymes based on genotype.41
| Phenotype | Genotype |
|---|---|
| UM | An individual with two increased function alleles or more than two normal function alleles (i.e., gene duplication). Generally considered to have increased enzyme activity compared with RMs. |
| RM | An individual with combinations of normal function and increased function alleles. Generally considered to have increased enzyme activity compared with NMs but less than UMs. |
| EM NM |
An individual with combinations of normal function and decreased function alleles (e.g., in the case of CYP2D6, the individual has at least one functional allele). Generally considered to have fully functional enzyme activity. |
| IM | An individual with combinations of normal function, decreased function, and/or no function alleles (e.g., in the case of CYP2D6, the individual has one decreased function and one no function allele). Generally considered to have decreased enzyme activity, between that of NMs and PMs. |
| PM | An individual with combination of no function alleles and/or decreased function alleles. Generally considered to have little to no enzyme activity. |
EM, extensive metabolizer; IM, intermediate metabolizer; NM, normal metabolizer; PM, poor metabolizer; RM, rapid metabolizer; UM, ultrarapid metabolizer.
REQUIRED RESOURCES
In order to effectively implement a clinical PGx service, there are a number of resources that are necessary prior to implementation. The following subsections can serve as an assessment tool to ensure that the appropriate evidence and resources are in place to facilitate establishment of a successful reactive clinical PGx program.
Evidence
In order to provide current evidence‐based, peer‐reviewed guidelines for the translation of pharmacogenetic test results into actionable clinical decisions, the CPIC was established in 2009 by the NIH‐funded Pharmacogenomics Research Network and Pharmacogenomics Knowledge Base.23 The CPIC guidelines are organized by the level of evidence available, which is divided into four tiers (A, B, C, and D). Levels A and B recommend at least one “change in prescribing” action, moderate/strong or optional, respectively, based on the patient's genotype.24 As of March 2016, the CPIC guidelines that contain level A or B evidence cover ∼17 genes and 87 drugs, although not all of the guidelines pertaining to these gene‐drug pairs have been published.25, 26 These guidelines are exceptional resources for determining the clinical relevance of gene‐drug pairs. Of note, the CPIC guidelines are intended to guide clinicians in understanding how to use available genetic test results to improve drug therapy, not to provide guidance on whether these tests should be ordered. Thoroughly disseminating this evidence among the different disciplines who prescribe the drug of interest in settings in which genetic information may apply at an institution is crucial to champion a clinical PGx program, particularly when seeking approval through the Pharmacy and Therapeutics, Medical Executive Committee, and for raising stakeholder and administrative support. Currently, few results from prospective PGx outcomes studies have been published, but several publications are in the pipeline, including those from PGx services at UF27 and UIC.28 Thus, it may be important to collect prospective data once service implementation is underway, including clinical outcomes (discussed in more detail later), to justify the benefit of your PGx service with regard to cost‐savings and safety/efficacy improvements at your institution.
Personnel
Building a strong interdisciplinary team is critical to ensuring successful implementation of a PGx service. An implementation team should consist of these stakeholders, at a minimum: PGx service leader, physician champion, clinician(s) who interpret genotyping test results and make recommendations, laboratory specialists, and educators. Clinicians with a background in PGx, medication management, and clinical pharmacology are ideally suited to lead and operate such a service. Particularly clinicians who have undergone residency, fellowship, or other training programs in PGx would be preferred. Many PGx programs described to date are led by pharmacists who work closely with other healthcare professionals to establish and run the program,11, 12, 15 whereas many PGx programs are also physician‐led.8, 9, 14, 16 If teaching is an institutional priority, a pharmacy or medical resident or fellow (with appropriate training and oversight) could assume many of the daily responsibilities of running the program, including documenting and interpreting genotype test results, under the direct supervision of clinical supervisor. Once the service clinician receives the genotype results from the laboratory and assesses the patient‐specific factors (including anthropometrics, overall clinical status, laboratory values, and drug‐drug interactions), he/she can make an informed drug therapy recommendation via a patient consult note within the EHR or directly to the provider. This note can then be electronically forwarded to all clinicians participating in the care of the patient. An alternative, and perhaps more practical and scalable, option to this drug therapy recommendation via a patient consult note or direct communication with the provider is to rely solely on clinical decision support (CDS) tools to facilitate genotype‐guided recommendations. More details on CDS will be discussed later.
Having a physician champion is imperative to PGx service success. Physician champions can advocate for gene‐drug pairs for which they feel that evidence supports implementation. Furthermore, they can provide support with regard to CDS creation, educate other physicians, and provide sponsorship in relation to administrative approval. Ideally, this physician champion should be highly regarded by his/her peers, personable, knowledgeable, invested, and autonomous. Having such a champion acting as an additional advocate will increase provider buy‐in and acceptance of PGx testing, which is a barrier that will be discussed in more detail in the “Challenges Faced with PGx Service Implementation” section. Good communication with the clinical staff is also important, so that any questions or concerns regarding genotype results or PGx recommendations can be voiced and addressed.
Information technology
To facilitate communication among the implementation team and ensure sharing of timely, easily accessible, and accurate PGx information among all, clinical PGx service implementation is only feasible at institutions with an EHR system. Having an informatics and information technology (IT) group at the institution will be fundamental to support alert creation (in the form of CDS, as detailed later) and for recording and integrating PGx test results into the EHR. Moreover, including a member on your implementation team with informatics/IT expertise can further facilitate this. The PGx tests are most useful if they can be easily ordered by clinicians and the results easily obtained. Unlike most clinical laboratory results, genotypes never change, so this information is pertinent throughout the patient's lifetime. Therefore, storing PGx test results in one designated location within the EHR can allow all involved healthcare practitioners to access and utilize this information over time. As with all parts of the PGx implementation program process, education is key in that clinicians, nurses, and other personnel must be informed where to find the genotype results and how to easily find the PGx service's contact information. It is imperative to keep in mind that building such areas within the EHR to house genotype results and creating CDS tools may take significant time to create. Thus, requests for builds within the EHR should be done early on in the planning stage. Some institutions may decide to include phenotype (e.g., poor metabolizer) and associated interpretations in plain language (e.g., absent enzyme activity) along with this genotype information.12
Laboratory
Understandably, access to laboratory services capable of genotyping variants of interest is a requirement. If the institution does not have a College of American Pathologists‐accredited/Clinical Laboratory Improvement Amendments (CLIA)‐licensed clinical laboratory on site and appropriately trained and licensed personnel, or if the laboratory cannot afford to rent or buy the appropriate genotyping platform(s), an outside laboratory can be used. Some academic medical centers or universities also offer outreach programs in which they will perform genotyping tests for other institutions. If an off‐site laboratory is used, there may be longer turnaround time from sample collection to genotype result(s), which may diminish the usefulness of genetic data, particularly when needed for urgent therapeutic decisions. The implementation team, nursing staff, and/or phlebotomy team must coordinate getting the patient's genetic sample to the laboratory in a timely manner; the EHR system should be able to facilitate this communication.
Examples of US Food and Drug Administration (FDA)‐cleared genotyping platforms are shown in Table 4. Alternatively, laboratory‐developed tests may be used for genotyping; although, whether the FDA will continue to allow laboratory‐developed tests is unclear. The benefit of using a platform cleared by the FDA is that the user can be assured that information on the platform's intended use, indications for use, methods, result interpretation, quality control and assay limitations, performance, clinical validity and interpretation, benefits and risks, and safety/efficacy have been extensively evaluated.38 Additionally, the FDA only regulates genotyping platforms that are sold as kits, which are groups of reagents used in genetic sample processing that are packaged together and sold to numerous laboratories. For those platforms that come to market as laboratory‐developed tests, meaning the test is developed and performed by a single laboratory and genetic samples are sent to that laboratory to be tested, the FDA has practiced “enforcement discretion.”39 We recommend that inexperienced PGx services use FDA‐cleared genotyping platforms, if possible.
Table 4.
| Gene | Trade name | Manufacturer | Allele variants detection | DNA source | Approximate turnaround time |
|---|---|---|---|---|---|
| CYP2D6 | xTAG CYP2D6 Kit v3 | Luminex Molecular Diagnostics | *1, *2, *3, *4, *5, *6, *7, *8, *9, *10, *11, *15, *17, *29, *35, *41, and duplication | Whole blood | 8–12 h |
| Roche AmpliChip CYP450 microarray | Roche Molecular Systems | *1, *2, *3, *4, *5, *6, *7, *8, *9,*10, *11, *15, *17, *19, *20, *29, *35, *36, *40, *41, *1XN (duplication), *2XN, *4XN, *10XN, *17XN, *35XN, *41XN | Whole blood | 6 h | |
| CYP2C19 | Spartan RX CYP2C19 Test System | Spartan Bioscience | *1,*2, *3, *17 | Buccal swab | 1 h |
| Verigene CYP2C19 Nucleic Acid Test | Nanosphere | *1, *2, *3, *17 | Whole blood | 2.5 h | |
| INFINITI CYP2C19 Assay | AutoGenomics | *1, *2, *3, *17 | Whole blood | 8 h44 | |
| Roche AmpliChip CYP450 microarray | Roche Molecular Systems, Inc. | *1, *2, *3, *17 | Whole blood | 6 h | |
| CYP2C9 and VKORC1 | eSensor warfarin sensitivity test and XT‐8 instrument | GenMark Diagnostics/ Osmetech Molecular Diagnostics | *2, *3, [*4, *5, *6, *11, *14, *15, *16],a and VKORC1 ‐1639 G>A |
|
3.5 h |
| eQ‐PCR LC warfarin genotyping kit | TrimGen Corporation | *2, *3, and VKORC1 ‐1639 G>A |
|
|
|
| Gentris Rapid Genotyping Assay ‐ CYP2C9 & VKORC1 | ParagonDx, LLC | *2, *3, and VKORC1 1173 C>T | Saliva | 1.5 h42 | |
| INFINITI 2C9 & VKORC1 Multiplex Assay for Warfarin | AutoGenomics | *2, *3, and VKORC1 ‐1639 G>A | Whole blood | 10.5 h42 | |
| Verigene Warfarin Metabolism Nucleic Acid Test and Verigene System | Nanosphere | *2, *3, and VKORC1 ‐1639 G>A | Whole blood | 1.5 h45 |
FDA, Food and Drug Administration.
These variants are part of an extended panel, which is not FDA‐cleared.
In addition to considering the FDA approval status of the genotyping platform, it would be wise to consider these additional factors when deciding upon a platform: cost, volume of expected samples, turnaround time of tests, time and labor required to run the test, and the number of genetic variants that can be tested for. Some manufacturers may allow institutions to borrow, lease, or buy the machine at a discounted price. This discounted price incentivizes the institution to buy the associated reagents and necessary materials for genotyping from that same company. Different platforms have previously been compared.40 The turnaround time of the genotyping test is a key factor that should weigh in on the institution's decision on which platform to use. If this turnaround time is too long, it may not be useful for clinical decision‐making regarding a patient's drug regimen. Furthermore, this turnaround time ties in with the time and labor of the laboratory personnel, which is an additional consideration when choosing a platform. Last, the number of variants that the platform can test for should be taken into account as well. For example, all of the various FDA‐cleared platforms relevant to warfarin pharmacogenetics test for a VKORC1 variant and the CYP2C9*2 and *3 allele, however, only a non‐FDA‐cleared extended panel tests for seven additional CYP2C9 variants that may also be clinically important, especially in persons of West African descent (Table 4).
Prior to initial use, the chosen instrument(s) and assays, regardless of whether they are FDA‐cleared or not, will need to be validated or verified according to quality assurance and control regulations (CLIA 42 CFR 493.1253 and College of American Pathologists GEN 42020‐42163). If these validation/verification processes are not performed, the laboratory may not release any test results. Along with the instrument calibration and calibration verification, the FDA‐cleared instrument must be tested for: (i) accuracy; (ii) precision; and (iii) reportable range of results for the test system. However, additional tests, including analytical sensitivity and specificity and reference intervals, are also required for non‐FDA‐cleared instruments. All test results must be documented and made readily available for CLIA certification survey, if needed. Requirement and timeframe for CLIA certification can differ by institution, therefore, it is highly recommended that the PGx team discuss these requirements with laboratory personnel and contact the CLIA State Agency or Regional Office for specific information. Information pertaining to CLIA certification can be found on the website for the Centers for Medicare & Medicaid Services (https://www.cms.gov/Regulations‐and‐Guidance/Legislation/CLIA/index.html). Furthermore, laboratory personnel will need to perform quality control every time samples are genotyped. It is recommended that laboratory personnel batch patient samples and run the genotype test(s) at most once daily. When to run the daily batch will depend on the approximate genotype result turnaround time for the institution's chosen platform (Table 4). Laboratory personnel must be thoroughly trained and licensed to use the genotyping platform to assure that results are accurate and timely, as well as to troubleshoot any problems that may arise. Often, the platform manufacturer will provide complementary training. Once genotyping results are available, the laboratory should efficiently alert the implementation team of the results and enter the results into the EHR.
PREPARATION FOR IMPLEMENTATION
In addition to gene‐drug pair selection and assessment of the resources required for PGx implementation, a PGx service implementation plan must be created to allow the stakeholders involved to understand the goals of the service and how they are to be achieved. Figure 1 shows a flowchart of some key steps involved in an implementation plan, which will be discussed in more detail in this section. A gene‐drug implementation process for the PGx service at UF has been previously published.12 Additionally, a plan should be formulated for data collection for quality assurance purposes at a minimum.
Figure 1.
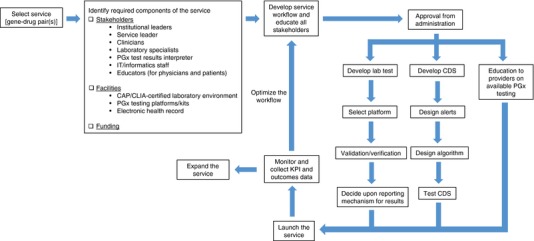
Reactive Pharmacogenomics Service Implementation Process. CAP, College of American Pathologists; CDS, Clinical decision support; CLIA, Clinical Laboratory Improvement Amendments; KPI, Key performance indicator; IT, Information Technology; PGx, Pharmacogenomics.
Data collection
Prior to implementing the PGx service, a plan should be made to determine what sort of data would be useful to collect. This includes key performance indicators (KPIs) as well as outcomes indicators. Collection of these service‐specific and patient‐specific variables is critical for quality assurance purposes, for cost‐analysis calculations, and for presenting to institutional administration to justify the benefit of the PGx service. It is important to note that, without approval from a human subject institutional review board these data would be for internal quality assurance/improvement purposes only and cannot be published.
Three major categories of KPIs for the PGx service are: (i) service process; (ii) service utilization; and (iii) patient and provider satisfaction. Example KPIs that could be collected for the PGx service are shown in Table 5. The KPIs focused on the service process allow the implementation team to document turnaround time of genotyping tests and quantify the overall efficiency of the current workflow. It is helpful to see how these indicators progress over time, and may allow the team to note areas within the workflow that may be improved upon. For instance, if the team notes that there is consistently a significant delay in the time it takes for the genotype order to result, further investigation may reveal an issue in the process of the blood sample being drawn and/or getting the sample to the laboratory once it is drawn. Perhaps there may be a delay in sample processing or genotyping once the laboratory receives the sample. This could reveal issues related to the assay, platform, or personnel efficiency. Next, KPIs concentrated on service utilization allow the team to observe the volume of PGx service consults by ordering medical service, by time frame, and as a ratio to see how frequently PGx testing for said drug is utilized compared with how often it is ordered. Last, KPIs can be a beneficial tool to gauge patient and provider satisfaction by survey or provider satisfaction that can be inferred by the providers’ overall PGx service recommendation acceptance rate. If available through the EHR, pilot testing using test patients should also be performed to help refine KPIs.
Table 5.
Example performance indicators for PGx service
| Category | Indicator |
|---|---|
| Service process |
|
| Service utilization |
|
| Satisfaction |
|
PGx, pharmacogenomics; EHR, electronic health record.
Besides collecting KPIs, it is fundamental to collect data on outcome indicators. These variables will certainly vary depending on the gene‐drug pair(s) that your institution decides to implement, and Table 6 contains some general examples, as well as potential comparison groups. Collecting outcome data may be more important in the near term, but perhaps not as important in the future once PGx testing becomes more widely accepted. For example, because of the current level of acceptance that TPMT or HLA‐B testing have achieved, collecting and reporting outcome data for these gene‐drug pairs may not be as impactful as it would be for other pairs that are less widely accepted. Data variables that may be beneficial to collect include hospitalizations associated with adverse drug effects of the implemented PGx drug or disease complications, such as stent thrombosis and other major cardiovascular events, if your institution were to implement CYP2C19 testing with clopidogrel.27 These outcomes could then be compared with historical control data at your institution to document improvement in outcomes with genotyping. Furthermore, outcomes indicator variables can be collected that relate to time to a therapeutic PGx drug dose (e.g., such as with genotype‐guided warfarin dosing) or drug level (e.g., such as with genotype‐guided tacrolimus dosing) and that relate to duration/use of a more expensive alternative drug. The latter is particularly important when calculating costs for cost‐effectiveness analyses. Many of these example outcome indicators can and should be used for such cost analyses, which will be necessary to assess the financial burden or benefit of the PGx service and justify continuation and/or possible expansion of the PGx service.
Table 6.
Examples of general outcomes indicator variables and comparison groups for data collection
| Outcomes indicator variables | Potential comparison groups |
|---|---|
|
|
ADE, adverse drug event; ED, emergency department; LOF, loss‐of‐function; PGx, pharmacogenomics.
Workflow development
PGx service workflow development is another essential process to assure service efficiency. PGx service workflow should cause minimal disruption to the clinicians’ workflow – therefore, involving clinicians in the design process will help develop a well‐accepted and effective service workflow. In most cases, completing the genotype test is the most time‐consuming step in the service workflow, and, thus, working with the testing laboratory to select a genotyping method that is maximally efficient can have significant impact on the overall PGx turnaround time. The PGx service workflow must include service‐related tasks and personnel/resources required for day‐to‐day service operation, including service coverage, hours of service operation, duration of patient follow‐up, and communication and documentation methods. In addition to facilitating service efficiency, having a good service workflow will also assist in the process of developing effective CDS tools. An example of the workflow for the warfarin PGx service at UIC has previously been published.15 Workflows will vary depending on the implemented gene‐drug pair, but most may include the following main elements: an automatic PGx service consult upon ordering a genotyping test; working‐up the consulted patient; writing a PGx service note in the EHR with PGx drug/dose recommendation after receiving genotyping test results; relaying the PGx information/recommendation to the appropriate team/clinicians; and storing the PGx information in the EHR. However, including all of these elements with implementation of every gene‐drug pair is likely not practical, especially for larger institutions. Thus, for larger and more sustainable implementation efforts, a well‐developed CDS can significantly streamline the workflow by providing essential information to guide the prescriber to order PGx testing and to assist with drug dosing/selection once genotype results are available.
Prior to implementation and periodically throughout, all relevant stakeholders should be educated on the purpose of the PGx service, its workflow, the location of genotype results in the EHR, and service contact information. This may be in the form of presentations at Grand Rounds, continuing education presentations, in‐services, and/or e‐learning. Moreover, education for providers can be facilitated via institution policies/procedures and a website created for the PGx service. This website can contain primary literature and summaries of the implemented gene‐drug pairs, including how the genetic testing information is utilized. Plus, a link for this website can be provided to clinicians via a CDS alert upon ordering a PGx drug. PGx service visibility could be increased with such measures as flyers and screensavers as well.
Administrative approval
In order to obtain participation from all necessary stakeholders, it is essential to obtain support from institutional leadership. Having support from all administrative levels, including health‐system administration, pharmacy, medicine, laboratory, and health IT, will help establish legitimacy for the service. At this stage, having a physician champion can have a large impact.
A common requirement for PGx services, particularly on the inpatient side, is approval by a regulatory body prior to implementation and periodic review to ensure that the program is of benefit. The Pharmacy and Therapeutics Committee generally fills the role of program approval and oversight; although additional approval from the Medical Executive Committee or other similar bodies may also be necessary in some settings.
Setup and testing of CDS
As implementation of CDS may affect the existing workflow and related personnel, developing an efficient CDS infrastructure is often considered one of the most challenging steps in the clinical PGx service implementation process. The CDS tools may be considered to: (i) notify or offer clinicians a genotyping test when a selected drug is ordered and notify the PGx service and laboratory personnel when a genotype test is ordered; (ii) provide concise and supportive information/guidance, a web‐link or contact information for additional assistance for clinicians prior to ordering the test; (iii) allow clinicians to easily complete the test orders within a short amount of time; and (iv) report the results in a way that can be easily viewed, alerting the implementation team and test orderer when the results become available. Once the genotyping results are available and stored within the EHR, point‐of‐care CDS should be created to alert clinicians of any prescribing implications.29
Additionally, certain CDS systems can be programmed to screen for patients eligible for genotyping by obtaining prespecified data from the EHR and automatically offering to order the genotyping test. CDS tools can also provide guidance or reference information at the time that the drug is ordered or generate an automatic smart documentation/order set pertinent to the patient's condition. For example, when UIC had automatic genotyping from August 2012 to April 2014 for patients who were newly initiated on warfarin, a CDS tool screened several places within the EHR to check for warfarin use in the past 6 months. When a prescriber ordered warfarin, the CDS tool automatically ordered the warfarin genotype test and PGx service consult. The CDS tool also provided a link that prescribers could click to read further information on available evidence for genotype‐guided warfarin dosing. Therefore, understanding the capability of the EHR and CDS should help the team to create an effective CDS interface. Nevertheless, there are situations when CDS might not be able to accurately perform the tasks programmed, such as when the required data for CDS are not available in the EHR, available data in the EHR are outdated, or additional assessment of the data is required. Given these important limitations, manual screening and assessment by PGx service personnel is sometimes still required to supervise CDS interventions.
To assist with incorporation of PGx information into EHRs with CDS, in 2013, the CPIC formed an Informatics Working Group. The initial focus of this group is to create comprehensive tables that translate genotype results to phenotype information to clinical recommendations for the CPIC guidelines.46 Newer CPIC guidelines contain such tables, sample CDS text, and figures that describe workflow from available result to CDS alert. For instance, the 2014 updated CPIC guidelines for HLA‐B Genotype and Abacavir Dosing contains an informatics section with this information.29 As an aside, it is imperative to keep the idea of alert fatigue in mind when designing your institution's CDS and workflow. In order to reduce the likelihood of encountering this phenomenon, it may be beneficial to only present alerts when it is necessary for clinicians to take action.29
After the initial CDS creation, extensive CDS system testing should be performed prior to the implementation to address possible errors that might occur. Additional information on CDS implementation process and support tools is available at the Agency for Healthcare Research and Quality (http://healthit.ahrq.gov/ahrq‐funded‐projects/clinical‐decision‐support‐cds‐initiative) and the Office of the National Coordinator for Health Information Technology websites (http://www.healthit.gov/providers‐professionals/clinical‐decision‐support‐cds).
STARTING IMPLEMENTATION
When all of the above elements are prepared and ready for implementation, a simultaneous process to monitor KPI data as well as service operations and workflow processes should also be initiated with the first genotype order. All issues that occur after service implementation should be documented, evaluated, and corrected within an appropriate timeframe (based on the urgency of the issues) to ensure workflow efficiency.
Communication with stakeholders
Frequent communication among primary team members is essential for the program to run efficiently. Specifically, the primary clinician covering the PGx service should communicate with the laboratory to ensure that genotyping is completed for the appropriate patients (i.e., patients for whom genotyping is indicated). The service clinician should also communicate with the medical team to ensure that appropriate medication recommendations are provided based on both genotype and clinical data. A pager and/or email and phone number specific to the PGx service will allow a centralized contact point for anyone requesting consultation from the service or if any questions arise.
Although the use of electronic CDS tools greatly facilitates communication among the implementation team members, and between the team and treating physicians, regular implementation team meetings are also important. These meetings allow the team to assess service performance (including KPI data), address workflow or personnel‐related issues, and communicate any changes in service procedures. Initially, it may be beneficial to have implementation team meetings weekly or monthly, adjusting the schedule according to necessity of discussions and frequency of encountered challenges. In addition, regular periodic meetings (for instance, quarterly) with institutional leadership may also be helpful to facilitate communication and allow discussions regarding existing program initiatives, potential future initiatives, and challenges that arise. At these meetings, outcomes and cost‐savings data are particularly relevant to discuss.
Last, communication with patients cared for by the service is highly recommended. Beyond simply reporting their genotype results, this often also includes educational materials that explain what genotyping is and why it was done. This is another critical element for a PGx service that is sometimes neglected. Therefore, provision of patient education materials for the particular gene‐drug pair must be included as part of the PGx service implementation plan. If satisfactory materials are not available for the chosen gene‐drug pair, then they will need to be developed in concert with clinicians and approved by institutional administration (the Pharmacogenomics Research Network and Pharmacogenomics Knowledge Base website can be a helpful resource). Furthermore, there must be a process in place to disseminate genotyping results and interpretations to patients (e.g., by mail or phone) in the event that they are discharged prior to finalization of the results.
Cost analysis
As with any service offered at most institutions, cost is a major factor. As such, it is likely that your team will need to create a marketing/business plan to justify the expenditures of the necessary personnel and equipment needed to implement a PGx service. It is crucial to highlight that while there are significant start‐up and maintenance costs involved in operating a PGx service, the savings may be well worth the expenditures. These expenditures include cost of the genotyping test, personnel, and genotyping platform, whereas the potential savings may include savings from avoiding adverse drug effects or treatment failure (Table 6). This underscores the importance of collecting outcomes data to demonstrate cost‐effectiveness of your PGx service.
It is prudent to present your PGx implementation plan to administration with regard to potential cost savings because your institution will likely be responsible for the incurred genotyping costs if your PGx service is implemented on the inpatient side. If the genotyping is performed on site in the institution's laboratory for an inpatient's clinical care, it is common for the laboratory to bill the hospital as part of a bundled fee for laboratory tests. Thus, your implementation team will need to discuss the entire plan with regard to cost expenditures and expected savings with key stakeholders. There is a paucity of cost‐effectiveness data based on clinical pharmacogenetic implementation data in support of PGx testing at the moment.47, 48 As PGx services may have the capacity to improve patients’ outcomes and save institutions money on readmissions due to adverse drug events, PGx implementation can make sense financially for the patient and the institution. On the other hand, outpatient testing is often directly billed to third‐party payers.
CHALLENGES FACED WITH PGx SERVICE IMPLEMENTATION
As with any new initiative at an institution, challenges are unavoidable, and implementing a PGx service is no exception to this. To address the challenges to clinical implementation of genomic medicine and support formation and exploration of practice models that aim to overcome these challenges in a multidisciplinary fashion, the National Human Genome Research Institute summoned investigators to create methods for, and assess feasibility of, integrating patients’ genomic results into their clinical care.18 Consequently, in 2013, the Implementing GeNomics In practice Network (www.ignite‐genomics.org) was formed. The Implementing GeNomics In practice Network is comprised of six genomic medicine demonstration projects that integrate genomic information into the EHR and offer CDS for implementation of clinically relevant interventions or advice across numerous and diverse sites.49 A main goal of Implementing GeNomics In practice is to develop best practices for clinical implementation of genomic medicine.49
Because challenges or barriers are inevitable and come up frequently, we discuss below several key challenges encountered by the PGx services at UF and UIC as well as ways these issues or barriers were addressed. Advanced awareness of such potential barriers may allow your implementation team to avoid or reduce the severity of future problems and design a workflow that takes some of these challenges into consideration. Five challenges that we will focus on are: (i) attaining provider buy‐in and acceptance of PGx testing; (ii) establishing genotyping and result interpretation; (iii) reimbursement for genetic testing; (iv) laboratory and workflow challenges; and (v) providing CDS to inform appropriate therapy.
Attaining provider buy‐in and acceptance of PGx testing
A potentially large hurdle to implementation of PGx can be gaining clinician acceptance of PGx testing. Early on during the planning stage, the implementation team must identify appropriate gene‐drug pairs with strong enough evidence pertaining to clinical utility. At UF, the decision was made to first implement the CYP2C19‐clopidogrel gene‐drug pair in patients undergoing left heart catheterization and percutaneous coronary intervention due to evidence that individuals carrying at least one loss‐of‐function CYP2C19 allele are at an increased risk of major adverse cardiovascular events.50, 51 Outcomes data collected by the UF implementation team in the first 2 years further corroborate this finding in their patient population, particularly in patients with acute coronary syndrome within 30 days post‐ percutaneous coronary intervention.27 This illustrates the value of collecting prospective outcomes data for your PGx service, which can be used to demonstrate the potential benefit of implementing a PGx service in your patient population.
Data collection in and of itself can present challenges as well. We have learned the importance of designing a data collection tool with the anticipated analyses already in mind. If data are collected without at least a basic analysis plan in place, significant time can be wasted cleaning and organizing the data into a format that will allow useful analysis. An argument often made is that there is not enough randomized controlled trial evidence available to support the benefit of genotype‐guided dosing for most gene‐drug pairs. Although that level of evidence is accepted as the gold standard for accessing a clinical intervention's efficacy, many laboratory tests currently used in clinical practice have not been validated with randomized controlled trials. Similar to these laboratory tests, PGx is one of many tools used to improve and individualize drug therapy.
Furthermore, we have experienced barriers with regard to clinician response to the PGx service's recommendations. With CYP2C9/VKORC1‐warfarin implementation at UIC, providers did not always accept our genotype‐guided warfarin dose recommendations for reasons such as they did not see our recommendation note in the EHR (e.g., they did not know to look for our note), they looked at the prior day's recommendation note, or they disagreed with our recommendation. If the PGx service does not already have a physician in its leadership, identifying a physician champion can be essential to overcoming a lack of provider buy‐in/acceptance of PGx service recommendations. As previously mentioned, physicians can provide tremendous support with regard to administrative approval, selection of appropriate gene‐drug pairs in a target patient subpopulation, CDS creation, and education of other clinicians.
We have also learned that data collection of both outcomes and KPIs, as well as effective education measures, are instrumental in addressing these barriers. As the PGx service is maintained over time, KPIs can be used to track the provider adherence rate to PGx service recommendations. Within the first 6 months of CYP2C9/VKORC1‐warfarin implementation at UIC, 73% of the daily warfarin doses ordered by the provider were within 0.5 mg of the dose recommendation by the PGx service.15 As of October 2015, this dose recommendation acceptance rate had increased to 85%. Data on KPIs and outcomes, once available, can also be included in clinician education, providing institution‐specific evidence of the PGx service's benefit, and may potentially lead to further increases in clinician acceptance of PGx service recommendations.
Educating clinicians and advertising the availability of such a service is critical to increase the exposure of the PGx service and the consult rate. Easily accessible CDS tools and check boxes for genotyping tests on order sets are also ways to improve the service's visibility and convenience of use. Increasing education regarding the PGx service workflow and the methods used to determine dose/drug recommendations may increase the adherence rate to PGx service recommendations. Expanding the visibility of the service and developing an effective CDS will be useful for such instances in which the provider did not see the PGx service's recommendation because he/she did not know to look for the note (for workflows similar to the model used for the warfarin PGx service at UIC). Over time, providers’ observance of PGx service recommendations and their effects on patients, in addition to providers building rapport with the PGx implementation team clinicians, can also be critical in overcoming barriers regarding provider acceptance. Additionally, as new evidence pertinent to your implemented gene‐drug pair(s) becomes available, the implementation team needs a plan to update relevant education materials, CDS tools, genotype result interpretations, and laboratory reports.
Establishing genotyping and result interpretation
In addition to the challenges we have faced with regard to getting support/acceptance from providers, we have also learned how crucial it is to consider the types of allelic variations that are clinically important within the selected gene when establishing genotyping. For instance, for any institutions deciding to implement a PGx service for drugs metabolized by CYP2D6, it is necessary to consider the complexities of the CYP2D6 locus when deciding upon which platform to use for genotyping. CYP2D6 is difficult to genotype due to its extremely polymorphic nature, including allelic variations, such as single nucleotide polymorphisms, gene copy number variation, insertions and deletions, and rearrangements with the related pseudogene CYP2D7.52 As such, if your institution were to implement CYP2D6, using the xTAG CYP2D6 Kit version 3 (Luminex, Austin, TX, USA) is a potential option due to this platform's flexibility, including the ability to capture gene duplications. However, when a genotype result from the xTAG CYP2D6 Kit version 3 platform is finalized, if there is gene duplication present (defined by Luminex as two or more gene copies per allele), it is difficult to determine the exact number of alleles present or which allele is duplicated. Thus, it may be challenging to determine a phenotype that represents the patients’ enzyme activity. Duplication of an allele with absent or reduced activity would produce a very different phenotype than a duplicated allele with normal activity. For example, a genotype result reported as CYP2D6*1/*41 duplication could result in an ultrarapid metabolizer phenotype if the patient's *1 allele (normal function) was duplicated, and an extensive (normal) metabolizer phenotype if the patient's *41 allele (reduced function) was duplicated (Table 3). In these unclear cases, the results could be reported using a phenotype range, for instance, extensive (normal) – ultrarapid metabolizer. However, phenotype prediction can be further complicated if the patient is on concomitant medications that can affect CYP2D6 activity, which, in some cases, can cause an extensive (normal) metabolizer to appear phenotypically as a poor metabolizer.
With CYP2C9/VKORC1‐warfarin implementation at UIC, the implementation team decided to use the eSensor XT‐8 System, as there is an extended panel that detects seven additional CYP2C9 variants that may have clinical relevance. This panel includes testing for CYP2C9*5, *6, and *11 alleles, which have been shown to significantly decrease warfarin clearance and dose requirements and are found nearly exclusively in individuals of West African descent.53, 54, 55, 56, 57, 58 This is particularly relevant to UIC's patient population, which is approximately half African American. Therefore, it is important to keep in mind your institution's patient population with consideration of the frequency of variation within the selected gene(s) when choosing a genotyping platform.
Reimbursement for genetic testing
Besides challenges related to choosing genotyping platforms and generating result interpretation, an evident challenge to all stakeholders is the challenge of obtaining reimbursement for these genotyping tests. This is one of the more emphasized barriers from an administrative perspective. At UF, the cost of CYP2C19 genotyping was covered by the implementation team and the pathology laboratory during the first year; in the first month after UF began billing third party payers, the reimbursement rate was 85% for outpatient claims.12 This is a significantly higher reimbursement rate than seen with CYP2C9/VKORC1 genotyping for warfarin dosing at UIC. Although PGx testing for warfarin may improve therapeutic dose predictions in a real world setting,28 PGx testing is considered investigational from the viewpoint of most health insurers, thus, most payers still do not provide coverage for it. However, some are willing to cover testing if the patient meets certain criteria, but with requirement of prior authorization for payment in most cases. For Medicare beneficiaries, genotype testing is only covered if it is ordered within 5 days of warfarin initiation to assist with dosing in warfarin‐naive patients who are enrolled in clinical trials that meet Medicare criteria.59 Reimbursement is a challenge that may not be easily overcome until more supportive evidence of clinical utility becomes available.
Laboratory and workflow challenges
In addition to reimbursement challenges, we have also encountered challenges related to the sample itself, including the type, timing, and quality of the sample. If your implementation team decides to implement a PGx service at outpatient sites, these sites may lack phlebotomy services. A potential solution is to use buccal cells rather than whole blood for PGx testing. Some drawbacks to this approach include the need to validate buccal cells for genotyping, as well as the added cost associated with buccal cell collection kits. If your implementation team has decided to utilize whole blood for DNA extraction, there can be barriers related to sample collection if all nurses and phlebotomists within the PGx service workflow are not properly educated or familiar with the genotype order. Both implementation teams at UF and UIC have faced challenges with this. This issue with sample collection was usually discovered upon evaluation of KPIs, because the turnaround time of the genotype test (from order to result) was substantially longer for some individuals. Upon further inquiry, we realized that some phlebotomists and nurses (those required to perform blood draws in the intensive care units) were not familiar with the genotype test order, did not know which tube to use, and/or did not know where to send the sample. Therefore, in some cases, the genotype test was not run or the test was delayed. Evaluation of KPIs and additional phlebotomy education has helped to overcome these issues.
With regard to processing samples for genotyping, the UIC implementation team has found it very useful to batch samples and only genotype samples once daily. We set a cutoff time of 9 am for running samples that day (any samples received after this would be genotyped the following day), allowing genotype results to be available prior to the second warfarin dose in most cases. When implementation first began, this cutoff time was set at 10 am and results would typically be back by 4 pm. With this earlier cutoff time, most genotype results are ready between 1 pm and 3:30 pm, as this change in the cutoff time allows quicker genotype turnaround time because it better suits the laboratory's current workflow. This earlier result time is especially important if the patient is found to have a warfarin‐sensitive genotype that would translate to a significantly lower dose recommendation than usual; in this case, the clinician covering the PGx service has more time to contact the prescriber to notify him/her of this result/recommendation before the end of the workday.
Providing CDS to inform appropriate therapy
Challenges related to sample type, collection, and processing are common at the outset of implementation, which may also be the case with the challenge of providing CDS to inform appropriate genotype‐guided therapy. As aforementioned, CDS is considered a vital part for PGx service implementation, however, it is also the part that must be customized to facilitate the current workflow the most; otherwise clinicians may be discouraged to order the genotype test.
Given the high prevalence of patients taking antidepressants, setting a CDS tool to alert providers with the suggestion to order CYP2D6 and CYP2C19 genotype tests may cause alert fatigue, because it is difficult to program a CDS tool to alert only with new medication orders. Therefore, the CDS may have to fire only when a test result already exists in the EHR. In addition, as evidence in the literature increases all the time, revision of genetic interpretations of the genotype results might be required at any time during or after PGx service implementation. Thus, these two issues must be considered before EHR and/or CDS tool procurement, and must be discussed with the IT team before implementing your PGx service to minimize burden to clinicians and maximize flexibility of the service to provide state of the art patient care.
At UIC, one major issue faced by the implementation team with the CDS tool is dealing with missing information in the EHR. The current warfarin dosing algorithm in use at UIC was designed to compute a suggested initial dose by using patients’ specific clinical factors from the EHR. A figure describing the CDS tool that generates this initial warfarin dose has been previously published.15 However, there are times that such information is not available in the EHR, and a suggested initial dose cannot be calculated and provided to clinicians within the alert. This issue could potentially raise hesitation for clinicians to order the genotype test as this absent dose recommendation may lead to confusion. Some of the common reasons behind missing information include: (i) the information is not obtained routinely or cannot be obtained; for example, baseline INR is not routinely checked if the patient is not taking warfarin or has liver disease; and (ii) the location of information in the EHR that is selected for the CDS tool might not be a common location used by clinicians/nurses for patients’ data entry. Hence, this issue must be addressed with the IT team during the CDS tool creation process and communicated with all stakeholders within the PGx workflow to ensure accuracy and consistency of the data entry process.
Several additional CDS‐related problems specific to your institution might also arise during the implementation process. In order to minimize the effect of these problems, extensive pilot testing of the CDS tool must be performed to identify any major issues that might affect service workflow.
CONCLUSION
Implementation of a reactive clinical PGx service is challenging but feasible. Accrued evidence supports the clinical utility of several gene‐drug pairs in routine practice. Strong PGx service leadership with good administrative support, a physician champion, an effective health IT system, College of American Pathologists‐accredited/ CLIA‐certified laboratory environment whether within or outside an institution, and well‐trained personnel are core elements for successful PGx service implementation. A systematic implementation plan with appropriate KPIs and outcome indicators should be created, monitored, and optimized to ensure service stability and efficiency prior to expanding the current PGx service. If there are adequate personnel and resources available to handle PGx service expansion, additional gene‐drug pairs can then be selected for implementation. Expanding the service often means additional workload and time commitment, and may require purchasing additional genotyping platforms if the current one does not have assays available for the gene(s) of interest. Additional administrative and financial support may also be needed. With the proper planning and resources, innovation through PGx services is achievable for hospital institutions nationwide.
Author Contributions
M.J.A. and S.C. performed background research and wrote the manuscript. L.H.C., E.A.N., and J.D.D. obtained the grant, performed background research, and reviewed/edited the manuscript.
Conflict of Interest
The authors declared no conflict of interest.
Acknowledgments
The authors would like to thank Alison Quinn, PharmD and Dyson Wake, PharmD for their thoughtful insight. L.H.C. is supported by NIH grant U01 HG007269; E.A.N. is supported by NIH grant K23 HL112908; J.D.D. is supported by NIH grant K23 GM112014. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Meghan J. Arwood and Supatat Chumnumwat contributed equally to this work.
References
- 1. Kalow, W. , Tang, B.K. & Endrenyi, L. Hypothesis: comparisons of inter‐ and intra‐individual variations can substitute for twin studies in drug research. Pharmacogenetics 8, 283–289 (1998). [DOI] [PubMed] [Google Scholar]
- 2. Johnson, J.A. & Cavallari, L.H. Pharmacogenetics and cardiovascular disease–implications for personalized medicine. Pharmacol. Rev. 65, 987–1009 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bosch, T.M. , Meijerman, I. , Beijnen, J.H. & Schellens, J.H. Genetic polymorphisms of drug‐metabolising enzymes and drug transporters in the chemotherapeutic treatment of cancer. Clin. Pharmacokinet. 45, 253–285 (2006). [DOI] [PubMed] [Google Scholar]
- 4. Kirchheiner, J. et al Pharmacogenetics of antidepressants and antipsychotics: the contribution of allelic variations to the phenotype of drug response. Mol. Psychiatry 9, 442–473 (2004). [DOI] [PubMed] [Google Scholar]
- 5. Patel, H.N. , Ursan, I.D. , Zueger, P.M. , Cavallari, L.H. & Pickard, A.S. Stakeholder views on pharmacogenomic testing. Pharmacotherapy 34, 151–165 (2014). [DOI] [PubMed] [Google Scholar]
- 6. Gottesman, O. et al The CLIPMERGE PGx Program: clinical implementation of personalized medicine through electronic health records and genomics‐pharmacogenomics. Clin. Pharmacol. Ther. 94, 214–217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Johnson, J.A. , Elsey, A.R. , Clare‐Salzler, M.J. , Nessl, D. , Conlon, M. & Nelson, D.R. . Institutional profile: University of Florida and Shands Hospital Personalized Medicine Program: clinical implementation of pharmacogenetics. Pharmacogenomics 14, 723–726 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. O'Donnell, P.H. et al The 1200 patients project: creating a new medical model system for clinical implementation of pharmacogenomics. Clin. Pharmacol. Ther. 92, 446–449 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pulley, J.M. et al Operational implementation of prospective genotyping for personalized medicine: the design of the Vanderbilt PREDICT project. Clin. Pharmacol. Ther. 92, 87–95 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shuldiner, A.R. et al Implementation of pharmacogenetics: the University of Maryland Personalized Anti‐platelet Pharmacogenetics Program. Am. J. Med. Genet. C Semin. Med. Genet. 166C, 76–84 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Crews, K.R. et al Development and implementation of a pharmacist‐managed clinical pharmacogenetics service. Am. J. Health Syst. Pharm. 68, 143–150 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Weitzel, K.W. et al Clinical pharmacogenetics implementation: approaches, successes, and challenges. Am. J. Med. Genet. C Semin. Med. Genet. 166C, 56–67 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hoffman, J.M. et al PG4KDS: a model for the clinical implementation of pre‐emptive pharmacogenetics. Am. J. Med. Genet. C Semin. Med. Genet. 166C, 45–55 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bielinski, S.J. et al Preemptive genotyping for personalized medicine: design of the right drug, right dose, right time‐using genomic data to individualize treatment protocol. Mayo Clin. Proc. 89, 25–33 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nutescu, E.A. et al Feasibility of implementing a comprehensive warfarin pharmacogenetics service. Pharmacotherapy 33, 1156–1164 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lee, J.A. et al Implementation and evaluation of a CYP2C19 genotype‐guided antiplatelet therapy algorithm in high‐risk coronary artery disease patients. Pharmacogenomics 16, 303–313 (2015). [DOI] [PubMed] [Google Scholar]
- 17. Levy, K.D. et al Prerequisites to implementing a pharmacogenomics program in a large health‐care system. Clin. Pharmacol. Ther. 96, 307–309 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Weitzel, K.W. et al The IGNITE network: a model for genomic medicine implementation and research. BMC Med. Genomics 9, 1 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Attia, J. et al How to use an article about genetic association: A: background concepts. JAMA 301, 74–81 (2009). [DOI] [PubMed] [Google Scholar]
- 20. Court, M.H. A pharmacogenomics primer. J. Clin. Pharmacol. 47, 1087–1103 (2007). [DOI] [PubMed] [Google Scholar]
- 21. Feero, W.G. , Guttmacher, A.E. & Collins, F.S. Genomic medicine–an updated primer. N. Engl. J. Med. 362, 2001–2011 (2010). [DOI] [PubMed] [Google Scholar]
- 22. Relling, M.V. & Evans, W.E. Pharmacogenomics in the clinic. Nature 526, 343–350 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Relling, M.V. & Klein, T.E. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin. Pharmacol. Ther. 89, 464–467 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. CPIC . Prioritization of CPIC genes/drugs diagram. [Internet]. 2016. https://cpicpgx.org/prioritization/#cpicLevels. Accessed 3 February 2016.
- 25. CPIC . Genes‐Drugs – CPIC. [Internet]. 2016. https://cpicpgx.org/genes‐drugs/. Accessed 3 February 2016.
- 26. Relling, M.V. , PGRN Session : Clinical Pharmacogenetics Implementation Consortium. [Presentation] presented at: ASCPT Annual Meeting; 9 Mar 2016; San Diego, CA: https://www.youtube.com/watch?v=uyaSfzvO4eA&feature=youtu.be. [Google Scholar]
- 27. Cavallari, L.H. et al Clinical implementation of CYP2C19‐genotype guided antiplatelet therapy reduces cardiovascular events after PCI [Abstract]. Circulation 132(Suppl 3), A11802 (2015). [Google Scholar]
- 28. Nutescu, E. et al Novel genotype guided personalized warfarin service improves outcomes in an ethnically diverse population [Abstract]. Circulation 130(Suppl 2), A16119 (2014). [Google Scholar]
- 29. Martin, M.A. et al Clinical Pharmacogenetics Implementation Consortium Guidelines for HLA‐B Genotype and Abacavir Dosing: 2014 update. Clin. Pharmacol. Ther. 95, 499–500 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Leckband, S.G. et al Clinical Pharmacogenetics Implementation Consortium guidelines for HLA‐B genotype and carbamazepine dosing. Clin. Pharmacol. Ther. 94, 324–328 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Scott, S.A. et al Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin. Pharmacol. Ther. 94, 317–323 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Crews, K.R. et al Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450 2D6 genotype and codeine therapy: 2014 update. Clin. Pharmacol. Ther. 95, 376–382 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Birdwell, K.A. et al Clinical Pharmacogenetics Implementation Consortium (CPIC) Guidelines for CYP3A5 Genotype and Tacrolimus Dosing. Clin. Pharmacol. Ther. 98, 19–24 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Relling, M.V. et al Clinical pharmacogenetics implementation consortium guidelines for thiopurine methyltransferase genotype and thiopurine dosing: 2013 update. Clin. Pharmacol. Ther. 93, 324–325 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Johnson, J.A. et al Clinical Pharmacogenetics Implementation Consortium Guidelines for CYP2C9 and VKORC1 genotypes and warfarin dosing. Clin. Pharmacol. Ther. 90, 625–629 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Suarez‐Kurtz, G . Pharmacogenomics in admixed populations. Trends Pharmacol. Sci. 26, 196–201 (2005). [DOI] [PubMed] [Google Scholar]
- 37. Scott, S.A. , Khasawneh, R. , Peter, I. , Kornreich, R. & Desnick, R.J. Combined CYP2C9, VKORC1 and CYP4F2 frequencies among racial and ethnic groups. Pharmacogenomics 11, 781–791 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. U.S. FDA . Guidance on pharmacogenetic tests and genetic tests for heritable markers [Internet]. 2016. http://www.fda.gov/RegulatoryInformation/Guidances/ucm077862.htm. Accessed 12 January 2016.
- 39. National Human Genome Research Institute . Regulation of Genetic Tests [Internet]. 2016. http://www.genome.gov/10002335. Accessed 12 January 2016.
- 40. Pratt, V.M. et al Characterization of 137 genomic DNA reference materials for 28 pharmacogenetic genes: a GeT‐RM Collaborative Project. J. Mol. Diagn. 18, 109–123 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pharmacogenomics Research Network and Pharmacogenomics Knowledge Base. CPIC Term Standardization for Clinical Pharmacogenetic Test Results Project [Internet]. 2015. https://www.pharmgkb.org/page/cpicTermProject. Accessed 4 February 2016.
- 42. Langley, M.R. , Booker, J.K. , Evans, J.P. , McLeod, H.L. & Weck, K.E. Validation of clinical testing for warfarin sensitivity: comparison of CYP2C9‐VKORC1 genotyping assays and warfarin‐dosing algorithms. J. Mol. Diagn. 11, 216–225 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. US Food and Drug Administration . Nucleic acid based tests [Internet]. 2015.. http://www.fda.gov/MedicalDevices/ProductsandMedicalProcedures/InVitroDiagnostics/ucm330711.htm. Accessed 25 July 2015.
- 44. Gladding, P. et al Pharmacogenetic testing for clopidogrel using the rapid INFINITI analyzer: a dose‐escalation study. JACC Cardiovasc. Interv. 2, 1095–1101 (2009). [DOI] [PubMed] [Google Scholar]
- 45. Lefferts, J.A. , Schwab, M.C. , Dandamudi, U.B. , Lee, H.K. , Lewis, L.D. & Tsongalis, G.J. Warfarin genotyping using three different platforms. Am. J. Transl. Res. 2, 441–446 (2010). [PMC free article] [PubMed] [Google Scholar]
- 46. Whirl‐Carrillo, M. , Hoffman, J.M. , Freimuth, R.R. & Peterson, J.F. TBI01: panel ‐ dissemination of pharmacogenomic knowledge: establishing a pathway to support clinical implementation. [Presentation] presented at: AMIA Joint Summits on Translational Science; 7 Apr 2014; San Francisco, CA. [Google Scholar]
- 47. Alagoz, O. , Durham, D. & Kasirajan, K. Cost‐effectiveness of one‐time genetic testing to minimize lifetime adverse drug reactions. Pharmacogenomics J. 16, 129–136 (2016). [DOI] [PubMed] [Google Scholar]
- 48. Schackman, B.R. , Haas, D.W. , Park, S.S. , Li, X.C. & Freedberg, K.A. Cost‐effectiveness of CYP2B6 genotyping to optimize efavirenz dosing in HIV clinical practice. Pharmacogenomics 16, 2007–2018 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. About IGNITE [Internet]. 2016.. https://www.ignite‐genomics.org/IGNITE_ABOUT.php. Accessed 24 February 2016.
- 50. Johnson, J.A. , Roden, D.M. , Lesko, L.J. , Ashley, E. , Klein, T.E. & Shuldiner, A.R. Clopidogrel: a case for indication‐specific pharmacogenetics. Clin. Pharmacol. Ther. 91, 774–776 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mega, J.L. et al Reduced‐function CYP2C19 genotype and risk of adverse clinical outcomes among patients treated with clopidogrel predominantly for PCI: a meta‐analysis. JAMA 304, 1821–1830 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gaedigk, A. Complexities of CYP2D6 gene analysis and interpretation. Int. Rev. Psychiatry 25, 534–553 (2013). [DOI] [PubMed] [Google Scholar]
- 53. Cavallari, L.H. et al Genetic and clinical predictors of warfarin dose requirements in African Americans. Clin. Pharmacol. Ther. 87, 459–464 (2010). [DOI] [PubMed] [Google Scholar]
- 54. Dickmann, L.J. et al Identification and functional characterization of a new CYP2C9 variant (CYP2C9*5) expressed among African Americans. Mol. Pharmacol. 60, 382–387 (2001). [DOI] [PubMed] [Google Scholar]
- 55. Drozda, K. et al Poor warfarin dose prediction with pharmacogenetic algorithms that exclude genotypes important for African Americans. Pharmacogenet. Genomics 25, 73–81 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Limdi, N.A. et al Influence of CYP2C9 and VKORC1 on warfarin dose, anticoagulation attainment and maintenance among European‐Americans and African‐Americans. Pharmacogenomics 9, 511–526 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Redman, A.R. , Dickmann, L.J. , Kidd, R.S. , Goldstein, J.A. , Ritchie, D.M. & Hon, Y.Y. CYP2C9 genetic polymorphisms and warfarin. Clin. Appl. Thromb. Hemost. 10, 149–154 (2004). [DOI] [PubMed] [Google Scholar]
- 58. Tai, G. et al In‐vitro and in‐vivo effects of the CYP2C9*11 polymorphism on warfarin metabolism and dose. Pharmacogenet. Genomics 15, 475–481 (2005). [DOI] [PubMed] [Google Scholar]
- 59. Centers for Medicare & Medicaid Services . National Coverage Determination (NCD) for Pharmacogenomic Testing for Warfarin Response (90.1) [Internet]. 2013. http://www.cms.gov/medicare‐coverage‐database/details/ncd‐details.aspx?NCDId=333&ncdver=1&bc=BAAAgAAAAAAA&. Accessed 23 July 2015. [PubMed]


