Abstract
Background
The clinical phenotype of Pseudoxanthoma elasticum (PXE) affected patients, although progressive with age, is very heterogeneous, even in the presence of identical ABCC6 mutations, thus suggesting the occurrence of modifier genes. Beside typical skin manifestations, the cardiovascular (CV) system, and especially the peripheral vasculature, is frequently and prematurely compromised.
Methods and results
A cohort of 119 Italian PXE patients has been characterized for apolipoprotein E (APOE) and methylenetetrahydrofolate reductase (MTHFR) gene polymorphisms by PCR. The severity of the clinical phenotype has been quantified according to the Phenodex PXE International score system. Statistical analysis (chi2 test, odd ratio, regression analysis, analysis of variance) were done by GraphPad. Data demonstrate that the frequency of APOE alleles is similar in PXE patients and in healthy subjects and that the allelic variant E2 confers a protection against the age-related increase of CV manifestations. By contrast, PXE patients are characterized by high frequency of the MTHFR-T677T polymorphism. With age, CV manifestations in T677T, but also in C677T, patients are more severe than those associated with the C677C genotype. Interestingly, compound heterozygosity for C677T and A1298C polymorphisms is present in 70% of PXE patients.
Conclusions
PXE patients may be screened for these polymorphisms in order to support clinicians for a better management of disease-associated CV complications.
Abbreviations: APOE, apolipoprotein E; CV, cardiovascular; Hcy, homocysteine; HDL, high density lipoproteins; MTHFR, methylenetetrahydrofolate reductase; PXE, Pseudoxanthoma elasticum
Keywords: Pseudoxanthoma elasticum, Skin-disease, Apolipoprotein E, Methylenetetrahydrofolate reductase, Cardiovascular
1. Introduction
Vascular calcification is a relevant clinical complication associated with aging, atherosclerosis, hypertension, chronic kidney disease and diabetes, being also correlated with increased risk of myocardial infarction. Furthermore, ectopic calcification occurs in a number of genetic diseases as in Pseudoxanthoma elasticum (PXE), a rare autosomal recessive disorder characterized by progressive mineralization of elastic fibers within soft connective tissues [1], mainly affecting skin, vascular walls and eyes [2]. In PXE, vessel alterations primarily consist of mineral plaques accumulating inside elastic fibers of medium sized arteries and veins. Therefore, the most common cardiovascular complications are: diminished or absent peripheral vascular pulses, mitral valve prolapse, arterial hypertension, angina pectoris, early intermittent claudication (often regarded as the first sign of accelerated atherosclerosis), arteriosclerosis and increased risk of myocardial and cerebral infarction [2], [3], [4], [5]. The age-dependent progression and severity of pathologic manifestations are very heterogeneous, thus suggesting that, beside ABCC6 causative mutations [6], [7], [8], other genes and/or polymorphisms contribute to the clinical phenotype [9], [10], [11], [12].
In addition to changes in the expression of molecules closely associated with the calcification process [13], [14], PXE subjects suffer from a condition of mild chronic oxidative stress [15], [16], [17]. It is well known that altered redox balance contributes to atherosclerosis and to vascular calcification and that several polymorphisms, as those in the apolipoprotein E (APOE) and 5,10 methylenetetrahydrofolate reductase (MTHFR) genes, are related to oxidative stress and to the development of cardiovascular complications [18].
In particular, APOE polymorphisms are associated with many diseases and it has been demonstrated that E2, E3 and E4 isoforms, having a different structure, exhibit a modified susceptibility to oxidation [19]. Consequently, APOE–protein interactions can be modified as well as protein function. APOE is one of the major lipid acceptor, removing cholesterol from cells and generating high density lipoprotein (HDL) particles in an isoform-dependent manner [20]. The increased risk of cardiovascular complications has been associated for instance to higher lipid levels in E4 carriers [21]. Moreover, it has been demonstrated that APOE isoforms, possibly depending on the number of free available –SH groups, have a different antioxidant activity (APOE2 > APOE3 > APOE4) [22], [23].
Another factor significantly contributing to oxidative damage in vascular-related diseases is homocysteine (Hcy), a non-protein amino acid capable of reacting specifically, and often quantitatively, with a number of thiol-combining groups, many of which are present in proteins and in other important molecules. Irreversible homocysteinylation of long-lived proteins lead to cumulative damages and to oxidative stress [24]. Hyperhomocysteinemia can be associated to MTHFR gene polymorphisms or to vitamin B6, B12 or folic acid deficiency and has been involved in the pathophysiology of cancer [25], [26], [27], cardio-cerebrovascular diseases, atherosclerosis [28], [29], renal failure [30] and diabetic retinopathy [31]. The MTHFR C677T polymorphism (rs1801133) has been extensively investigated and the corresponding amino acid substitution results in a 30% to 60% decrease of the enzyme activity in heterozygotes and homozygotes, respectively [32]. A second common mutation within the MTHFR gene, A1298C (rs1801131), results in significantly reduced enzyme catalytic activity, even though only few studies have focused on this polymorphism [33], [34]. It has been also suggested that the two polymorphisms (i.e. compound heterozygosity for C677T and A1298C) presumably act in an additive manner increasing the risk, for example, for ischemic stroke [35].
In the present study Italian PXE patients have been investigated: 1) for the frequency of APOE allelic variants and of MTHFR C677T polymorphism in order to evaluate if they are randomly present in patients or if there is an association with the disease; 2) to see if the severity of cardiovascular manifestations are influenced by these polymorphisms, thus representing additive genetic risk factors that could be counteracted by appropriate life-style.
2. Materials and methods
2.1. Patient's clinical data
Analyses were performed on 119 Pseudoxanthoma elasticum (PXE) affected patients and on 103 healthy volunteers. Since PXE is more frequently observed in females than in males [2], we have maintained a female:male ratio of 2:1 within each group.
All subjects were of Italian origin and gave informed signed consent in accordance with the guidelines of the Institutional Medical Ethical Committee and of the Helsinki Declaration of 1975 revised in 1983.
Clinical diagnosis of PXE was confirmed by the presence of calcified elastic fibers on skin biopsies and on the identification of ABCC6 causative mutations (Supplemental Table 1) detected by already described methods [36]. Age-matched healthy individuals were selected on the basis of the absence of clinically evident pathologic conditions and of cardiovascular complications.
The severity of the pathological phenotype, as documented by clinical examinations, has been quantified according to the Phenodex score that represents, nowadays, the only published standardized system quantifying PXE clinical manifestations [37].
In particular, for the vascular system, V1 was given to weak or absent pulses, V2 to intermittent claudication and V3 to vascular occlusion or other symptoms severe enough to require surgery. For cardiac symptoms, C1 denoted ischemic changes and angina, and C2 myocardial infarction. The cardiovascular score (CV) used in the present study was the sum of V + C.
Cholesterol, high density lipoproteins (HDL), low density lipoproteins (LDL), triglycerides and homocysteine were measured in PXE patients by chemical analyses in certified laboratories under the control of the Local Sanitary Units. Patients under statin therapy were analyzed for polymorphisms, but their laboratory tests were not considered.
2.2. Identification of APOE polymorphisms
Genomic DNA was isolated from whole blood (QIAamp blood kit, Qiagen GmbH, Hilden, Germany), according to the manufacturer's instructions and stored at − 80 °C until use.
Samples underwent the polymerase chain reaction (PCR) protocol described by Hixson and Vernier [38]. Briefly, DNA was amplified using oligonucleotide forward primer (5’-ACAGAATTCGCCCCGGCCTGGTACACAC-3’) and reverse primer (5’-TAAGCTTGGCACGGCTGTCCAAGGA-3’) (MWG-Biotech, Ebersberg, Germany) [39].
In a total volume of 30 μl, each amplification reaction contained 0.3 μg of genomic DNA from each patient, 3 μl of PCR Buffer 10x (20 mM Tris–HCl pH 7.5, 100 mM KCl, 15 mM MgCl2, 1 mM DTT, 0.1 mM EDTA, 0.5% Tween 20, 0.5% Nonidet P40, 50% glycerol) (Roche, Milan, Italy), 3 μl of each primer (MWG-Biotech), 0.6 μl of dNTPs mix (10 mM of dATP, dCTP, dGTP, dTTP) (Fermentas, Milan, Italy), 3 μl (10%) dimethyl sulfoxide (DMSO), 0.5 μl of High Fidelity Taq DNA polymerase corresponding to 1.73 U (Roche) plus sterile water. Samples were heated at 94 °C for 5 min for denaturation, and subjected to 30 cycles of amplification by primer annealing (6O°C for 1 min), extension (70 °C for 2 min.) and denaturation (95 °C for 1 min) in a T3 Thermocycler (Biometra, Goettingen, Germany).
PCR amplification products were evaluated by agarose gel electrophoresis. Samples were digested by adding 1 μl of the restriction enzyme HhaI (10 U/μl, Fermentas) for 3 h at 37 °C. Each reaction mixture was loaded onto an agarose gel Metaphor (Cambrex Bio Science, Walkersville, USA) 4% in TAE 1 × allowing to discriminate PCR products with small size variations. Electrophoresis was performed for 1 h and 30 min at 80 V. DNA fragments were visualized by UV illumination and the size of HhaI fragments was estimated by comparison with 0.1 μg of Gene Ruler 100 bp DNA Ladder Plus (Fermentas).
2.3. Identification of MTHFR polymorphisms
Detection of C677T MTHFR polymorphism was performed by PCR followed by HinfI restriction enzyme digestion. Briefly, we used the forward primer 5′-CCT TGA ACA GGT GGA GGC CAG-3′ and the reverse primer 5′-GCG GTG AGA GTG GGG TGG AG-3′ (Invitrogen, Monza, Italy) to amplify a 294 base pair (bp) fragment of the MTHFR gene. Each 25 μl PCR reaction contained 2.5 μl of 10x reaction buffer (Roche), 1.5 mmol MgCl2 (Roche), 2 μl from 10 pmol of each primer, 0.2 mmol of the deoxynucleoside triphosphates, 1 U of Taq DNA polymerase (Roche), and 100 ng of genomic DNA template. The mixture was denatured at 95 °C for 5 min, and the PCR reaction was performed for 35 cycles in a thermocycler under the following conditions: denaturation at 95 °C for 1 min, annealing at 65 °C for 30 s, and extension at 72 °C for 1 min. The final extension cycle of 72 °C was for 7 min. The PCR products were electrophoresed on agarose gel (2%) to confirm the correct amplicon size. Restriction enzyme digestion was performed on PCR products using the HinfI restriction enzyme (Fermentas) following the supplier's protocol. After digestion, all fragments were resolved on a metaphor agarose gel (5%) (BIOSPA, Milan, Italy). A single fragment of 294 bp and two fragments of 168 and 126 bp identified the homozygous (CC) and (TT) genotype, respectively, whereas three fragments of 294, 168, and 126 bp were indicative of the heterozygous (CT) condition.
The A1298C polymorphism was detected by PCR in a total volume of 50 μl, containing: 5 μl of 10x reaction buffer (Roche), 10 pmol of the forward primer 5′-CTT TGG GGA GCT GAA GGA CTA CTA C-3′ and 10 pmol of the reverse primer 5′-CAC TTT GTG ACC ATT CCG GTT TG-3′, 10 mM of dNTP mix, 3.0 mM MgCl2 (Roche) and 1 unit Taq polymerase (Roche) and 200 ng of genomic DNA. PCR parameters were as follows: an initial denaturation step of 2 min at 92 °C, followed by 35 cycles of 92 °C for 60 s, 51 °C for 60 s, 72 °C for 30 s and a final extension for 7 min at 72 °C to ensure a complete extension of all PCR products. PCR products were electrophoresed on an agarose gel (2%) to confirm the correct amplicon size (163 bp). The sequence analysis of this PCR fragment was performed by automated sequencing (ABI Prism, model 377, version 2.1.2, Monza, Italy), using the ABI Prism Taq DyeDeoxy terminator cycle sequencing ready reaction kit (Perkin Elmer, Waltham, USA), according to the manufacturer's instructions.
2.4. Statistical analysis
GraphPad version 4.0 software was used for statistical analyses. Differences between subgroups were tested by ANOVA; categorical variables were tested by χ2 analysis or by Fisher's exact test. Deviation of genotype frequencies from those predicted by Hardy–Weinberg law was tested by χ2 analysis. Odds ratios (OR) and 95% confidence intervals (CI) were calculated to estimate the relative risk of the different genotype combinations. Significance was set at p < 0.05.
3. Results and discussion
3.1. Frequency of APOE and MTHFR polymorphisms
All observed frequencies fitted with those expected according to the Hardy–Weinberg equilibrium.
The APOE genotype and the allele frequency distribution in PXE and in healthy subjects are reported in Table 1. The frequencies in the control group were similar to those already reported for the general population [40]. No significant differences between PXE and healthy subjects were observed with regard to either genotype or allele frequencies. As expected [41], the E3E3 genotype was the most common in both groups, E2E2 was absent in both controls and patients, whereas E4E4 was absent in patients, but present in 1.9% of healthy subjects and E2E4 was present in 1.7% and 1% of patients and healthy subjects, respectively.
Table 1.
Allele and genotype frequencies of APOE polymorphisms in control and in PXE subjects.
There were no statistically significant differences between PXE patients and control subjects.
| Genotype/allele | Control (%) (n = 103) | PXE (%) (n = 119) | OR (95% CI) | p valuea |
|---|---|---|---|---|
| Genotype | ||||
| E2E2 | 0 | 0 | ||
| E3E3 | 63.1 | 74.1 | 1.00 (reference) | |
| E4E4 | 1.9 | 0 | 0.1705 (0.008 –3.619) | 0.22 |
| E2E3 | 12.6 | 10.3 | 0.6549 (0.2688–1.595) | 0.37 |
| E3E4 | 21.4 | 13.8 | 0.5676 (0.2667–1.208) | 0.18 |
| E2E4 | 1.0 | 1.7 | 1.703 (0.1507–19.23) | 1.0 |
| Allele | ||||
| e2 | 7 | 6 | 0.7973 (0.2569–2.474) | 0.78 |
| e3 | 80 | 86 | 1.00 (reference) | |
| e4 | 13 | 8 | 0.5725 (0.2254–1.454) | 0.25 |
Fisher's test was used; p < 0.05 was considered to be statistically significant.
In contrast to APOE, several studies indicate that the frequency of the homozygosity for the MTHFR-C677T polymorphism varies significantly (from 1.4% to 15%) depending on the geographic area [42]. Consistently, the risk of stroke associated with the T677T genotype is significantly elevated among the Japanese and the Italian population [28]. Therefore, we have evaluated genotype and allele frequency of the C677T variant in PXE patients and in control subjects (Table 2). Odds ratio indicates that the relative frequencies of genotypes differ between PXE and control subjects (p < 0.05). In particular, in healthy controls, it was observed that the frequency of CC and TT genotypes were 34.0% and 17.5%, respectively. To be noted is that the percentage of control individuals with the TT polymorphism is higher than that reported in population from other countries [43], but in agreement with data from Sacchi et al. [44]. In PXE, the CC and the TT genotypes account for 24.3% and 27.7% of patients, while 48% were heterozygous for the CT genotype. Interestingly, the homozygous genotype MTHFR T677T had a higher frequency in PXE patients (27.7%) than in controls (17.5%) (Table 2).
Table 2.
Allele and genotype frequencies of the MTHFR (C677T) polymorphism in controls and in PXE patients. The frequency of MTHFR T677T genotype was higher in PXE patients than in the control group.
| Genotype/allele | Control (%) (n = 103) | PXE (%) (n = 119) | OR (95% CI) | p valuea |
|---|---|---|---|---|
| Genotype | ||||
| CC | 34.0 | 24.3 | 1.00 (reference) | |
| CT | 48.5 | 48.0 | 1.417 (0.7335–2.736) | 0.32 |
| TT | 17.5 | 27.7 | 2.333 (1.051–5.182) | 0.04 |
| Allele | ||||
| C | 58 | 48 | 1.00 (reference) | |
| T | 42 | 52 | 1.496 (0.8560–2.615) | 0.16 |
Fisher's test was used; p < 0.05 was considered to be statistically significant (values in bold).
3.2. APOE polymorphisms and cardiovascular manifestations
Due to their low frequency, subjects with the ε2/ε4 genotype (1.7%) were excluded and were not considered in future analyses. PXE patients were divided into three APOE subgroups: subjects carrying the ε2/ε3 genotype (APOE2 group), the ε3/ε3 genotype (APOE3 group) and the ε3/ε4 genotype (APOE4 group). As expected [45], total cholesterol and low density lipoprotein levels progressively increased with age in APOE3 and APOE4 patients, whereas changes were not significant in APOE2 subjects. Plasma triglyceride levels were not modified by age in any APOE subgroups (Supplemental Fig. 1). Epidemiological data in the general population have demonstrated that the APOE genotype is associated with increased risk of cardiovascular disease [46]. Therefore, we have assessed if also in PXE APOE genotype had an influence on the severity of cardiovascular manifestations. Since in PXE CV complications usually start at 30 years and then progress with age, patients, divided according to their APOE genotype, were further stratified into 3 age groups: < 30 years; 30–50 years and > 50 years.
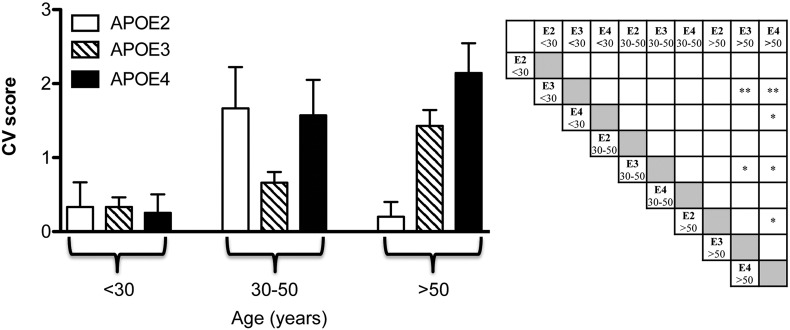
As shown in Fig. 1, PXE patients over the fifth decade and with APOE3 or APOE4 genotype had a CV score higher than those with the same genotype in the lowest age group (< 30 years). Moreover, in older patients (> 50 years) with the APOE4 genotype, CV complications were more severe than those in APOE2 subjects. These results demonstrate that in PXE, as well as in the general population [46], APOE2 confers protection against age-related CV manifestations.
Fig. 1.
The cardiovascular score (CV) in PXE patients according to the APOE genotype (APOE2: E2E3; APOE3: E3E3 and APOE4: E3E4). Patients were divided into three age subgroups (before 30 years of age, between the age of 30 and 50 years, after the age of 50 years). Significant p values are indicated in the right panel as stars. *p < 0.05; **p < 0.01.
3.3. MTHFR polymorphisms and cardiovascular manifestations
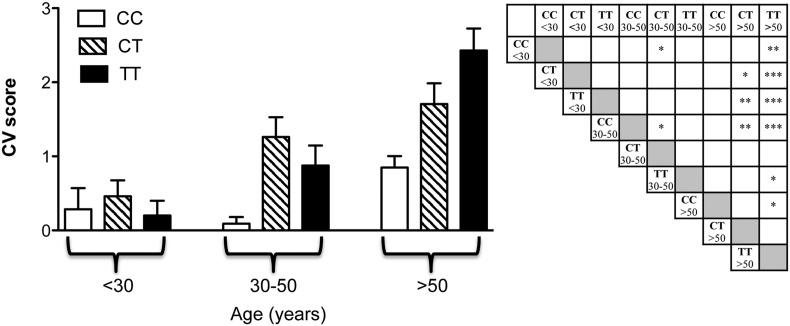
Similar analyses have been done for the MTHFR genotype (Fig. 2), one of the factors that favor increased Hcy levels. Hcy can induce vascular injuries [47], extracellular matrix alterations [48] and connective tissues structural remodeling [49]. Fig. 2 shows the CV score as a function of both MTHFR genotype and patients' age. The CV score increased with age with significant differences in patients with C677T or T677T genotype. Consistently, PXE patients with the C677C genotype did not exhibit a significant age-dependent increase of the CV score, whereas PXE subjects with C677T or T677T polymorphism presented, after 30 years of age, a continuous progression in the severity of CV manifestations. In order to better understand why the CV score was similarly affected in the CT heterozygous and in the TT homozygous condition, we have investigated the occurrence of polymorphism A1298C in patients heterozygous for C677T. It has in fact been demonstrated that the A1298C polymorphism can affect MTHFR activity and Hcy concentration [51], especially if combined with the C677T variant, thus leading to a condition similar to that observed in patients carrying the T677T genotype. Interestingly, 70% of PXE patients was compound heterozygous for both MTHFR polymorphisms (C677T and A1298C), thus behaving similarly to patients with the T677T polymorphism.
Fig. 2.
The cardiovascular score (CV) in PXE patients according to the MTHFR genotype (CC: C677C; CT: C677T and TT: T677T). Patients were divided into three age subgroups (before 30 years of age, between the age of 30 and 50 years, after the age of 50 years). Significant p values are indicated in the right panel as stars. *p < 0.05; **p < 0.01; ***p < 0.001.
Genetic alterations of enzymes involved in Hcy metabolism or inadequate supply of folate, vitamin B12 or vitamin B6 lead to elevated levels of Hcy. Laboratory tests indicate that, in PXE patients, there is also a graded increase in plasma Hcy concentrations from the CC to TT genotypes of MTHFR–C677T polymorphism (CC = 9.4 mg/dl; CT = 15.5 mg/dl; TT = 21.2 mg/dl). Within this context, it is worth mentioning that in PXE patients high Hcy levels can be efficiently reverted to normal values by folate supplementation (e.g. 43.03 μmol/l at basal levels were reduced to 8.57 μmol/l by vitamin B6 intake), thus demonstrating that the disease does not affect patients' response to vitamin treatment.
However, since plasma Hcy levels are rapidly modified by diet and/or vitamin supplementation, in terms of prognostic significance, genetic analysis, being independent from these variables, represents the most useful parameter to be investigated.
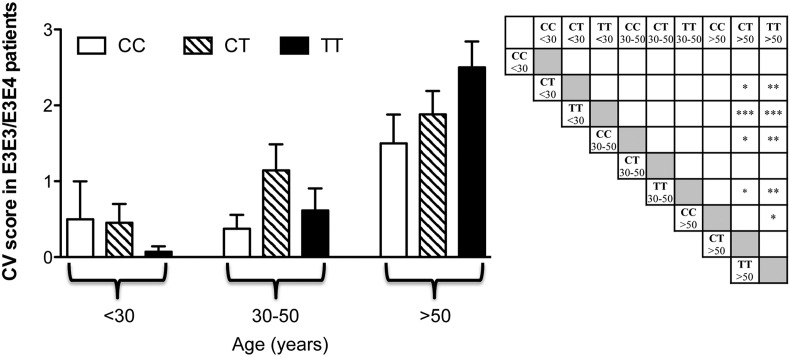
APOE and Hcy are risk factors for cardiovascular diseases and can influence each other's activities [50]. For instance, Hcy impairs APOE3 function by interfering with the dimerization of APOE3 and reducing the APOE3-mediated HDL generation to a level similar to that of APOE4, although it does not affect E4 isoform. Since PXE patients with the APOE3 or the APOE4 genotype behave similarly, in contrast to those with the APOE2 genotype, we have further analyzed patients with the APOE3 or the APOE4 genotype, dividing these subjects into subgroups on the basis of age and MTHFR–C677T polymorphism. Fig. 3 shows that the CV score, as expected, increased with age, however, despite the low number of patients present in each group, the CV score significantly increased in patients with C677T or T677T polymorphisms over the fifth decade of life compared to younger patients. Therefore, these data indicate that MTHFR polymorphisms can further modulate the severity of cardiovascular manifestations in PXE patients.
Fig. 3.
The cardiovascular score (CV) was evaluated in APOE3 and APOE4 PXE patients stratified according to MTHFR polymorphisms (CC: C677C; CT: C677T and TT: T677T) and age. In particular, patients were divided into three age subgroups (before 30 years of age, between the age of 30 and 50 years and after the age of 50 years). Significant p values are indicated in the right panel as stars. *p < 0.05; **p < 0.01; ***p < 0.001.
4. Conclusion
Polymorphisms in the APOE and MTHFR genes are known to be important risk factors for cardiovascular diseases. Since vascular manifestations represent one of the most life-threatening complications in PXE patients, the identification of prognostic markers could be of great importance.
Results from the present study demonstrate that: a) the frequency of APOE polymorphisms is similar between patients and healthy subjects, indicating the absence of a link between PXE and a specific APOE genotype; b) as known for the general population, also in PXE patients, the ε3 and ε4 alleles are associated with a similarly higher age-dependent increase of cardiovascular manifestations compared to ε2 patients. Moreover, as far as the MTHFR gene it has been observed that: a) T677T polymorphism has been found at a frequency higher in patients than in controls; b) with age, PXE patients homozygous for the C677C polymorphism do not exhibit a significant age-dependent increase of the CV score as patients carrying C677T and T677T polymorphisms; c) compound heterozygosity for C677T and A1298C polymorphisms is present in 70% of PXE patients; d) MTHFR polymorphisms influence the age-dependent increase of the CV score in PXE patients further modulating the APOE genotype.
Although this study is limited to a number slightly over one hundred of patients, it has to be underlined that, since PXE is a rare disease, this represents a consistent cohort, that, most importantly, is of homogeneous geographical origin. Ethnic differences must therefore be considered when the frequency of specific mutations/polymorphisms is evaluated in larger groups of patients. Data from this study suggest that MTHFR polymorphisms may be playing a role in PXE cardiovascular complications and, since Hcy levels can be controlled by appropriate vitamin supplementation, PXE patients could be investigated for both MHTFR polymorphisms in order to adopt appropriate diet regimens to counteract the “at-risk” MTHFR genotype.
Competing interests
No conflict of interests are disclosed.
Acknowledgments
Authors would like to thank the PXE patients for their collaboration to this study. The work is supported by PXE Italia Onlus.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ymgmr.2014.11.002.
Appendix A. Supplementary data
Supplementary material.
References
- 1.Gheduzzi D., Sammarco R., Quaglino D., Bercovitch L., Terry S., Taylor W., Ronchetti I.P. Extracutaneous ultrastructural alterations in pseudoxanthoma elasticum. Ultrastruct. Pathol. 2003;27:375–384. [PubMed] [Google Scholar]
- 2.Quaglino D., Boraldi F., Annovi G., Ronchetti I. The multifaceted complexity of genetic diseases: a lesson from pseudoxanthoma elasticum. In: Ikehara K., editor. Advances in the Study of Genetic Disorders. InTech; Rijeka, Croazia: 2011. pp. 289–318. [Google Scholar]
- 3.Lefthériotis G., Omarjee L., Le Saux O., Henrion D., Abraham P., Prunier F., Willoteaux S., Martin L. The vascular phenotype in pseudoxanthoma elasticum and related disorders: contribution of a genetic disease to the understanding of vascular calcification. Front. Genet. Feb 12 2013;4:4. doi: 10.3389/fgene.2013.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lebwohl M.G., Distefano D., Prioleau P.G., Uram M., Yannuzzi L.A., Fleischmajer R. Pseudoxanthoma elasticum and mitral-valve prolapse. N. Engl. J. Med. 1982;307:228–231. doi: 10.1056/NEJM198207223070406. [DOI] [PubMed] [Google Scholar]
- 5.Challenor V.F., Conway N., Monro J.L. The surgical treatment of restrictive cardiomyopathy in pseudoxanthoma elasticum. Br. Heart J. 1988;59:266–269. doi: 10.1136/hrt.59.2.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Le-Saux O., Urban Z., Tschuch C., Csiszar K., Bacchelli B., Quaglino D., Pasquali-Ronchetti I., Pope F.M., Richards A., Terry S., Bercovitch L., de-Paepe A., Boyd C.D. Mutations in a gene encoding an ABC transporter cause pseudoxanthoma elasticum. Nat. Genet. 2000;25:223–227. doi: 10.1038/76102. [DOI] [PubMed] [Google Scholar]
- 7.Bergen A.A., Plomp A.S., Schuurman E.J., Terry S., Breuning M., Dauwerse H. Mutations in ABCC6 cause pseudoxanthoma elasticum. Nat. Genet. 2000;25:228–231. doi: 10.1038/76109. [DOI] [PubMed] [Google Scholar]
- 8.Ringpfeil F., Lebwohl M.G., Christiano A.M., Swart J., Kool M., van Soest S., Baas F., ten Brink J.B., de Jong P.T. Pseudoxanthoma elasticum: mutations in the MRP6 gene encoding a transmembrane ATP-binding cassette (ABC) transporter. Proc. Natl. Acad. Sci. U. S. A. 2000;97:6001–6006. doi: 10.1073/pnas.100041297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hendig D., Langmann T., Kocken S., Zarbock R., Szliska C., Schmitz G., Kleesiek K., Götting C. Gene expression profiling of ABC transporters in dermal fibroblasts of pseudoxanthoma elasticum patients identifies new candidates involved in PXE pathogenesis. Lab. Invest. 2008;88:1303–1315. doi: 10.1038/labinvest.2008.96. [DOI] [PubMed] [Google Scholar]
- 10.Zarbock R., Hendig D., Szliska C., Kleesiek K., Götting C. Pseudoxanthoma elasticum: genetic variations in antioxidant genes are risk factors for early disease onset. Clin. Chem. 2007;53:1734–1740. doi: 10.1373/clinchem.2007.088211. [DOI] [PubMed] [Google Scholar]
- 11.Zarbock R., Hendig D., Szliska C., Kleesiek K., Götting C. Vascular endothelial growth factor gene polymorphisms as prognostic markers for ocular manifestations in pseudoxanthoma elasticum. Hum. Mol. Genet. 2009;18:3344–3351. doi: 10.1093/hmg/ddp259. [DOI] [PubMed] [Google Scholar]
- 12.Zarbock R., Hendig D., Szliska C., Kleesiek K., Götting C. Analysis of MMP2 promoter polymorphisms in patients with pseudoxanthoma elasticum. Clin. Chim. Acta. 2010;411:1487–1490. doi: 10.1016/j.cca.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 13.Gheduzzi D., Boraldi F., Annovi G., Paolinelli-DeVincenzi C., Schurgers L.J., Vermeer C., Quaglino D., Pasquali-Ronchetti I. Matrix gla protein is involved in elastic fiber calcification in the dermis of pseudoxanthoma elasticum patients. Lab. Invest. 2007;87:998–1008. doi: 10.1038/labinvest.3700667. [DOI] [PubMed] [Google Scholar]
- 14.Boraldi F., Annovi G., Vermeer C., Schurgers L.J., Trenti T., Tiozzo R., Guerra D., Quaglino D. Matrix gla protein and alkaline phosphatase are differently modulated in human dermal fibroblasts from PXE patients and controls. J. Invest. Dermatol. 2013;133:946–954. doi: 10.1038/jid.2012.460. [DOI] [PubMed] [Google Scholar]
- 15.Pasquali-Ronchetti I., Garcia-Fernandez M.I., Boraldi F., Quaglino D., Gheduzzi D., Paolinelli-DeVincenzi C., Tiozzo R., Bergamini S., Ceccarelli D., Muscatello U. Oxidative stress in fibroblasts from patients with pseudoxanthoma elasticum: possible role in the pathogenesis of clinical manifestations. J. Pathol. 2006;208:54–61. doi: 10.1002/path.1867. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Fernandez M.I., Gheduzzi D., Boraldi F., Paolinelli C.D., Sanchez P., Valdivielso P., Morilla M.J., Quaglino D., Guerra D., Casolari S., Bercovitch L., Pasquali-Ronchetti I. Parameters of oxidative stress are present in the circulation of PXE patients. Biochim. Biophys. Acta. 2008;1782:474–481. doi: 10.1016/j.bbadis.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 17.Boraldi F., Annovi G., Guerra D., Paolinelli Devincenzi C., Garcia-Fernandez M.I., Panico F., De Santis G., Tiozzo R., Ronchetti I., Quaglino D. Fibroblast protein profile analysis highlights the role of oxidative stress and vitamin K recycling in the pathogenesis of pseudoxanthoma elasticum. Proteomics Clin. Appl. 2009;3:1084–1098. doi: 10.1002/prca.200900007. [DOI] [PubMed] [Google Scholar]
- 18.Xin X.Y., Song Y.Y., Ma J.F., Fan C.N., Ding J.Q., Yang G.Y., Chen S.D. Gene polymorphisms and risk of adult early-onset ischemic stroke: a meta-analysis. Thromb. Res. 2009;124:619–624. doi: 10.1016/j.thromres.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 19.Jolivalt C., Leininger-Muller B., Bertrand P., Herber R., Christen Y., Siest G. Differential oxidation of apolipoprotein E isoforms and interaction with phospholipids. Free Radic. Biol. Med. 2000;28:129–140. doi: 10.1016/s0891-5849(99)00232-4. [DOI] [PubMed] [Google Scholar]
- 20.Kolovou G.D., Anagnostopoulou K.K. Apolipoprotein E polymorphism, age and coronary heart disease. Ageing Res. Rev. 2007;6:94–108. doi: 10.1016/j.arr.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 21.Zende P.D., Bankar M.P., Kamble P.S., Momin A.A. Apolipoprotein E gene polymorphism and its effect on plasma lipids in arteriosclerosis. J. Clin. Diagn. Res. 2013;7:2149–2152. doi: 10.7860/JCDR/2013/6195.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyata M., Smith J.D. Apolipoprotein E allele-specific antioxidant activity and effects on cytotoxicity by oxidative insults and beta-amyloid peptides. Nat. Genet. 1996;14:55–61. doi: 10.1038/ng0996-55. [DOI] [PubMed] [Google Scholar]
- 23.Pedersen W.A., Chan S.L., Mattson M.P. A mechanism for the neuroprotective effect of apolipoprotein E: isoform-specific modification by the lipid peroxidation product 4-hydroxynonenal. J. Neurochem. 2000;74:1426–1433. doi: 10.1046/j.1471-4159.2000.0741426.x. [DOI] [PubMed] [Google Scholar]
- 24.Starkebaum G., Harlan J.M. Endothelial cell injury due to copper-catalyzed hydrogen peroxide generation from homocysteine. J. Clin. Invest. 1986;77:1370–1376. doi: 10.1172/JCI112442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frosst P., Blom H.J., Milos R., Goyette P., Sheppard C.A., Matthews R.G., Boers G.J., den Heijer M., Kluijtmans L.A., van den Heuvel L.P., Rozen R. A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nat. Genet. 1995;10:111–113. doi: 10.1038/ng0595-111. [DOI] [PubMed] [Google Scholar]
- 26.Hamid A., Wani N.A., Kaur J. New perspectives on folate transport in relation to alcoholism-induced folate malabsorption—association with epigenome stability and cancer development. FEBS J. 2009;276:2175–2191. doi: 10.1111/j.1742-4658.2009.06959.x. [DOI] [PubMed] [Google Scholar]
- 27.Yin G., Ming H., Zheng X., Xuan Y., Liang J., Jin X. Methylenetetrahydrofolate reductase C677T gene polymorphism and colorectal cancer risk: a case-control study. Oncol. Lett. 2012;4:365–369. doi: 10.3892/ol.2012.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cronin S., Furie K.L., Kelly P.J. Dose-related association of MTHFR 677 T allele with risk of ischemic stroke: evidence from a cumulative meta-analysis. Stroke. 2005;36:1581–1587. doi: 10.1161/01.STR.0000169946.31639.af. [DOI] [PubMed] [Google Scholar]
- 29.Trabetti E. Homocysteine, MTHFR gene polymorphisms, and cardio-cerebrovascular risk. J. Appl. Genet. 2008;49:267–282. doi: 10.1007/BF03195624. [DOI] [PubMed] [Google Scholar]
- 30.Födinger M., Sunder-Plassmann G. Methylenetetrahydrofolate reductase polymorphisms and renal failure. In: Ueland P.M., Rozen R., editors. MTHFR Polymorphisms and Disease. Eurekah.com/Landes Bioscience; Georgetown,Texas: 2005. pp. 170–178. [Google Scholar]
- 31.Yigit S., Karakus N., Inanir A. Association of MTHFR gene C677T mutation with diabetic peripheral neuropathy and diabetic retinopathy. Mol. Vis. 2013;19:1626–1630. [PMC free article] [PubMed] [Google Scholar]
- 32.Rozen R. Genetic predisposition to hyperhomocysteinemia: deficiency of methylenetetrahydrofolate reductase (MTHFR) Thromb. Haemost. 1997;78:523–526. [PubMed] [Google Scholar]
- 33.Weisberg I., Tran P., Christensen B., Sibani S., Rozen R. A second genetic polymorphism in methylenetetrahydrofolate reductase (MTHFR) associated with decreased enzyme activity. Mol. Genet. Metab. 1998;64:169–172. doi: 10.1006/mgme.1998.2714. [DOI] [PubMed] [Google Scholar]
- 34.Sazci A., Ergul E., Tuncer N., Akpinar G., Kara I. Methylenetetrahydrofolate reductase gene polymorphisms are associated with ischemic and hemorrhagic stroke: dual effect of MTHFR polymorphisms C677T and A1298C. Brain Res. Bull. 2006;71:45–50. doi: 10.1016/j.brainresbull.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 35.Han I.B., Kim O.J., Ahn J.Y., Oh D., Hong S.P., Huh R., Chung S.S., Kim N.K. Association of methylenetetrahydrofolate reductase (MTHFR 677C>T and 1298A>C) polymorphisms and haplotypes with silent brain infarction and homocysteine levels in a Korean population. Yonsei Med. J. 2010;51:253–260. doi: 10.3349/ymj.2010.51.2.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gheduzzi D., Guidetti R., Anzivino C., Tarugi P., DiLeo E., Quaglino D., Pasquali Ronchetti I. ABCC6 mutations in Italian families affected by pseudoxanthoma elasticum. Hum. Mutat. 2004;24:438–439. doi: 10.1002/humu.9284. [DOI] [PubMed] [Google Scholar]
- 37.Pfendner E.G., Vanakker O.M., Terry S.F., Vourthis S., McAndrew P.E., McClain M.R., Fratta S., Marais A.S., Hariri S., Coucke P.J., Ramsay M., Viljoen D., Terry P.F., De Paepe A., Uitto J., Bercovitch L.G. Mutation detection in the ABCC6 gene and genotype–phenotype analysis in a large international case series affected by pseudoxanthoma elasticum. J. Med. Genet. 2007;44:621–628. doi: 10.1136/jmg.2007.051094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hixson J.E., Vernier D.T. Restriction isotyping of human apolipoprotein E by gene amplification and cleavage with HhaI. J. Lipid Res. 1990;31:545–548. [PubMed] [Google Scholar]
- 39.Emi M., Wu L.L., Robertson M.A., Myers R.L., Hegele R.A., Williams R.R., White R., Lalouel J.M. Genotyping and sequence analysis of apolipoprotein E isoforms. Genomics. 1988;3:373–379. doi: 10.1016/0888-7543(88)90130-9. [DOI] [PubMed] [Google Scholar]
- 40.Corbo R.M., Scacchi R., Mureddu L., Mulas G., Alfano G., Apolipoprotein E. Polymorphism in Italy investigated in native plasma by a simple polyacrylamide gel isoelectric focusing technique. Comparison with frequency data of other European populations. Ann. Hum. Genet. 1995;59:197–209. doi: 10.1111/j.1469-1809.1995.tb00741.x. [DOI] [PubMed] [Google Scholar]
- 41.Brouwer D.A., Doormaal J.J., vanMuskiet F.A. Clinical chemistry of common apolipoprotein E isoforms. J. Chromatogr. B Biomed. Appl. 1996;678:23–41. doi: 10.1016/0378-4347(95)00256-1. [DOI] [PubMed] [Google Scholar]
- 42.Motulsky A. Nutritional ecogenetics: homocysteine-related arteriosclerotic vascular disease, neural tube defects, and folic acid. Am. J. Hum. Genet. 1996;58:17–20. [PMC free article] [PubMed] [Google Scholar]
- 43.Klujitmans L.A.J., van den Heuvel L.P.W.J., Boers G.H.J., Frosst P., Stevens E.M.B., van Oost B.A., den Heijer M., Trijbles F.J.M., Rozen R., Blom H.J. Molecular genetic analysis in mild hyperhomocysteinemia: a common mutation in the methylene-tetrahydrofolate reductase gene is a genetic risk factor for cardiovascular disease. Am. J. Hum. Genet. 1996;58:35–41. [PMC free article] [PubMed] [Google Scholar]
- 44.Sacchi E., Tagliabue L., Duca F., Mannucci P.M. High frequency of the C677T mutation in the methylenetetrahydrofolate reductase (MTHFR) gene in Northern Italy. Thromb. Haemost. 1997;78:963–964. [PubMed] [Google Scholar]
- 45.Connor S.L., Connor W.E., Sexton G., Calvin L., Bacon S. The effects of age, body weight and family relationships on plasma lipoproteins and lipids in men, women and children of randomly selected families. Circulation. 1982;65:1290–1298. doi: 10.1161/01.cir.65.7.1290. [DOI] [PubMed] [Google Scholar]
- 46.Haan M.N., Mayeda E.R. Apolipoprotein E genotype and cardiovascular diseases in the elderly. Curr. Cardiovasc. Risk Rep. 2010;4:361–368. doi: 10.1007/s12170-010-0118-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCully K.S. Chemical pathology of homocysteine. I. Atherogenesis. Ann. Clin. Lab. Sci. 1993;23:477–493. [PubMed] [Google Scholar]
- 48.Tyagi S.C. Homocysteine redox receptor and regulation of extracellular matrix components in vascular cells. Am. J. Physiol. 1998;274:C396–C405. doi: 10.1152/ajpcell.1998.274.2.C396. [DOI] [PubMed] [Google Scholar]
- 49.Mujumdar V.S., Tummalapalli C.M., Aru G.M., Tyagi S.C. Mechanism of constrictive vascular remodeling by homocysteine: role of PPAR. Am. J. Physiol. Cell Physiol. 2002;282:1009–1015. doi: 10.1152/ajpcell.00353.2001. [DOI] [PubMed] [Google Scholar]
- 50.Minagawa H., Watanabe A., Akatsu H., Adachi K., Ohtsuka C., Terayama Y., Hosono T., Takahashi S., Wakita H., Jung C.G., Komano H., Michikawa M. Homocysteine, another risk factor for Alzheimer disease, impairs apolipoprotein E3 function. J. Biol. Chem. 2010;285:38382–38388. doi: 10.1074/jbc.M110.146258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van der Put N.M., Gabreëls F., Stevens E.M., Smeitink J.A., Trijbels F.J., Eskes T.K., van den Heuvel L.P., Blom H.J. A second common mutation in the methylenetetrahydrofolate reductase gene: an additional risk factor for neural-tube defects? Am. J. Hum. Genet. 1998;62:1044–1051. doi: 10.1086/301825. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.