Abstract
The diagnosis of invasive fungal infections (IFIs) is usually based on the isolation of the fungus in culture and histopathological techniques. However, these methods have many limitations often delaying the definitive diagnosis. In recent years, molecular diagnostics methods have emerged as a suitable alternative for IFI diagnosis. When there is not a clear suspicion of the fungus involved in the IFI, panfungal real-time PCR assays have been used, allowing amplification of any fungal DNA. However, this approach requires subsequent amplicon sequencing to identify the fungal species involved, increasing response time. In this work, a new panfungal real-time PCR assay using the combination of an intercalating dye and sequence-specific probes was developed. After DNA amplification, a melting curve analysis was also performed. The technique was standardized by using 11 different fungal species and validated in 60 clinical samples from patients with proven and probable IFI. A melting curve database was constructed by collecting those melting curves obtained from fungal species included in the standardization assay. Results showed high reproducibility (coefficient of variation [CV] < 5%; r > 0.95) and specificity (100%). The overall sensitivity of the technique was 83.3%, with the group of fungi involved in the infection detected in 77.8% of the positive samples with IFIs covered by molecular beacon probes. Moreover, sequencing was avoided in 67.8% of these “probe-positive” results, enabling report of a positive result in 24 h. This technique is fast, sensitive, and specific and promises to be useful for improving early diagnosis of IFIs.
INTRODUCTION
Invasive fungal infections (IFIs) remain a major cause of morbidity and mortality in immunocompromised patients, and diagnosis continues to be problematic (1). Laboratory diagnosis of IFIs is based on classical methods such as fungus isolation in culture and histopathological examination; however, these methods present several limitations. Fungal cultures are frequently slow growing, and such assays are low in sensitivity as well as in specificity because fungi are habitual laboratory contaminants and part of the saprophytic human flora (2). Furthermore, histopathological studies show low sensitivity, require skilled personnel, and do not distinguish among fungal species (3), which is problematic since such distinctions are crucial in order to define an appropriate antifungal therapy, due to the differences in antifungal susceptibility exhibited by different fungal species (4).
In recent years, molecular methods, such as PCR assays, have emerged as a suitable alternative to conventional methods for the diagnosis of IFIs. These assays have a higher sensitivity as they allow the detection of small amounts of DNA in clinical samples. In addition, those protocols based on quantitative real-time PCR (qPCR) have the benefit of quantifying the fungal burden in clinical specimens (5). Several qPCR protocols have been developed for the diagnosis of IFIs, mainly for Aspergillus and Candida species (6) but also for less frequent fungal species (7, 8, 9, 10). However, the utility of a species or genus-specific approach is limited when there is not a clear suspicion of the fungus involved in the IFI. To solve this limitation, panfungal or broad-range fungal PCR assays have been described as an appropriate alternative. However, these techniques also present the inconvenience of the requirement of sequencing after amplification, which involves a delay in definite diagnosis. To date, several studies based on panfungal PCR showing suitable results (11, 12, 13) and demonstrating its usefulness in certain groups of patients, such as hemato-oncology patients (14) and immunocompromised pediatric patients (15), have been described.
Melting curve analysis has been reported by several authors as a fast, reliable, and cost-effective method to identify fungal species while avoiding sequencing (16, 17, 18, 19, 20). Moreover, a combination of probe detection and high-resolution melting analysis has been already used for detecting and identifying Aspergillus spp. from clinical samples (21). However, to our knowledge, this is the first method developed for IFI diagnosis that combines panfungal and species-specific detection with species identification by using melting curve analysis in the same run.
In this work, a panfungal qPCR assay combining a DNA binding dye and specific molecular beacon probes followed by a melting curve analysis has been designed in order to detect a wide range of fungal species and decrease response time by avoiding sequencing as far as possible. The technique has been standardized and validated for clinical strains and clinical samples from patients with proven and probable IFI. The aim of this study was to improve early diagnosis of IFIs in patients when there is not a clear suspicion of the fungus involved in the infection.
MATERIALS AND METHODS
Control strains.
All strains included in the assay belonged to the fungal collection of the Spanish National Centre for Microbiology. DNA from the following organisms (strains in parentheses) was used to standardize the new panfungal qPCR assay: Rhizopus oryzae (CNM-CM 3020), Rhizopus microsporus (CNM-CM 4244), Mucor circinelloides (CNM-CM 2437), Aspergillus flavus (CNM-CM 3509), Aspergillus terreus (CNM-CM 3508), Aspergillus fumigatus (CNM-CM 2580), Histoplasma capsulatum var. capsulatum (CNM-CM 2721), Histoplasma capsulatum var. duboisii (CNM-CM 4626), Coccidioides immitis (CNM-CM 7056), Coccidioides posadasii (CNM-CM 2911), Paracococcidioides brasiliensis (CNM-CM 2908), and Blastomyces dermatitidis (CNM-CM 3114).
To assess the specificity of the technique, the following yeast and mold strains were used: Scedosporium apiospermum (CNM-CM 3169), Scedosporium prolificans (CNM-CM 1627), Fusarium solani (CNM-CM 3530), Fusarium oxysporum (CNM-CM 3197), Cunninghamella elegans (CNM-CM 7046), Penicillium commune (CNM-CM 7192), Cryptococcus gattii (CNM-CL 5007), Cryptococcus neoformans (CNM-CL 5801), Candida albicans (CNM-CL 8701), Candida krusei (CNM-CL 7057), Candida parapsilosis (CNM-CL 5683), Candida glabrata (CNM-CL 7523), Candida tropicalis (CNM-CL 8796), Candida guilliermondii (CNM-CL 7127), Rhizomucor pusillus (CNM-CM 2751), Actinomucor elegans (CNM-CM 1722), Lichtheimia corymbifera (CNM-CM 7053), and Lichtheimia ramosa (CNM-CM 7130). With the aim of evaluating the detection of Aspergillus spp. and Mucor spp., the following strains were also included: Aspergillus lentulus (CNM-CM 6069), Aspergillus niger (CNM-CM 4352), Aspergillus viridinutans (CNM-CM 5623), Neosartorya udagawae (CNM-CM 6056), Neosartorya pseudofischeri (CNM-CM 2270), Aspergillus novofumigatus (CNM-CM 6098), Aspergillus ustus (CNM-CM 4212) Mucor plumbeus (CNM-CM 5245), Mucor velutinosus (CNM-CM 6560), and Mucor irregularis (CNM-CM 7301).
Primer and probe design.
Primers used in the assay were the universal primer ITS1 and a modified universal primer, ITS2 (22), which was redesigned with two degenerated positions to avoid deviation in the amplification efficiency among species (ITS2Deg2). Five molecular beacon probes labeled with different fluorescent dyes were designed to specifically target the ITS1 region of the rDNA from different groups of fungal species (Aspergillus spp., Mucorales, and fungi involved in endemic mycoses) based on a large database of internal transcribed spacer (ITS) sequences (containing more than 10,000 distinct sequences). Beacon Designer 7.0 software (Premier Biosoft, Palo Alto, CA, USA) was used for probe design. The primers and probes designed were subjected to a BLAST search within the GenBank sequence database (http://blast.ncbi.nlm.nih.gov/Blast.cgi) to avoid cross-homology with other microorganisms. Sequences of primers and probes used in this assay are shown in Table 1.
TABLE 1.
Sequences of primers and probes designed for the new panfungal qPCR assaya
| Primer or probeb | Sequence |
|---|---|
| Primers | |
| ITS1 (f) | 5′-TCCGTAGGTGAACCTGCGG-3′ |
| ITS2Deg2 (r) | 5′-GCTRCGTTCTTCATCGATRC-3′ |
| Probes | |
| PanAspDeg | 5′-ROX-CGCGATCAACCTCCCACCCGTGWCTAYYGTACCGATCGCG-BHQ2-3′ |
| PanRhiz | 5′-HEX-CGCGATTTCTGGGGTTTGATCGATGCCAATCGCG-BHQ1-3′ |
| PanMuc | 5′-HEX-CGCGATCGGCTTGGTATCCTATTATTATTTACCAAAAAGAATT GATCGCG-BHQ1-3′ |
| PanHBP | 5′-Cy5-CGCGATTCGGCGGGCCTGCAGCGATCGCG-BHQ2-3′ |
| PanCocci3 | 5′-Cy5-CGCGATCGCGCCTGCCAGYGGATCAATTATCGCG-BHQ2-3′ |
f, forward; r, reverse; BHQ1, black hole quencher 1; BHQ2, black hole quencher 2. Degenerated positions are in bold, and stem regions are underlined.
Primers were used to amplify the ITS1 region from the fungal rDNA. Molecular beacon-specific probes were designed to detect each group of fungi, as follows: MB-PanAspDeg, Aspergillus species; MB-PanRhiz, Rhizopus species; MB-PanMuc, Mucor species; MB-PanHBP, Histoplasma, Blastomyces, and Paracoccidioides species; MB-PanCocci3, Coccidioides species.
Panfungal real-time PCR assay.
The amplification assay was carried out in an LC480 unit (Roche Diagnostics, Mannheim, Germany). PCRs were performed in a 20-μl final volume containing 2× SensiMix II Probe No-ROX (Bioline, Ecogen, Madrid, Spain), 0.8 μM each primer, 0.2 μM MB-PanAspDeg, 0.2 μM MB-PanMuc, 0.1 μM MB-PanRhiz, 0.3 μM MB-PanHBP, 0.3 μM MB-PanCocci3, and 1× Resolight binding dye (Roche Diagnostics, Mannheim, Germany) diluted 16 times. Finally, 2 μl of genomic DNA from control strains or 2 or 4 μl of DNA extracted from samples was added to PCRs in standardization and validation experiments, respectively. PCR conditions were as follows: an initial step of 10 min at 95°C, followed by 50 cycles at 95°C for 10 s, 54°C for 30 s, and 72°C for 30 s and a cooling phase of 30 s at 40°C. Melting curves were generated by increasing the temperature from 65°C to 99°C at 0.29°C/s. Results were considered positive when the fluorescent signal above the baseline was detected as determined by second-derivative analysis and were expressed in terms of the quantification cycle (Cq). Each experiment included quantification standards of five species of fungi as well as negative controls. Subsequently, a color compensation experiment was performed to prevent cross talk between dyes.
Standardization.
Standard curves for the fungal species mentioned above in “Control strains” were obtained based on the result of five PCR repetitions with 10-fold serial dilutions of genomic DNA ranging from 1 ng to 0.1 fg/μl of reaction mixture. Two replicates of each dilution were included in each PCR repetition to evaluate the intrareproducibility of the technique. Regression lines were obtained by plotting the logarithm of the initial template concentration versus the corresponding Cq, and the standard curve was then used to determine the sensitivity, primer efficiencies, and reproducibility of the assay. In addition, the coefficient of variation (CV) was determined in each case.
To evaluate the specificity of the technique, 0.1 ng/μl of genomic DNA from 18 fungal species as well as mouse and human DNA (Promega, Madrid, Spain) was included in duplicate in the PCR assay.
Melting curve database construction.
Melting curves of each PCR product were obtained by measuring the fluorescence of the Resolight Dye at different temperatures (from 65°C to 99°C) and analyzed by using the melting curve genotyping analysis included in LightCycler 480 Software v. 1.5 (Roche Diagnostics, Mannheim, Germany). Melting curves were automatically displayed as a melting curve chart (fluorescence [F] versus temperature [Ti]) and plotted by representing the first negative derivative melting curve (−df/dt) versus temperature, where the center of the melting peak corresponded to the point of inflection of the melting curve. This point of inflection represents the melting temperature of the PCR product. A melting curve database was generated by collecting, as external melting standards, those melting curves generated for each species included in the standardization assay presenting the best quality.
Validation assay in clinical samples.
The usefulness of the new panfungal qPCR assay was evaluated using 60 clinical samples. Thirty-seven of the 60 clinical samples belonged to patients with proven IFI, and the remaining 23 belonged to patients with probable IFI, classified according to the EORTC/MSG criteria (23). Of the IFI samples, 17 samples belonged to patients with invasive aspergillosis, 11 with mucormycosis, 10 with infection by endemic mycoses, 9 with candidiasis, and 13 with IFIs caused by emerging or rare fungal species (Table 2). The origins of the samples were as follows: 44 were biopsy specimens (35 being fresh and the remaining paraffin-embedded biopsy specimens), 8 were respiratory samples (5 bronchoalveolar lavage fluids, 2 bronchoaspiration specimens, and 1 tracheal aspirate), 5 were cerebrospinal fluids (CSF), 1 was aqueous humor fluid, and 1 was a nail sample. DNA extraction from clinical samples was performed manually by using a QIAamp DNA minikit (Qiagen, Hilden, Germany) according to the manufacturer's instructions and as already described by Buitrago et al. (24) without any preprocessing step. Biopsy samples embedded in paraffin were deparaffinized by lavage with 1.5 ml of xylene (100%) followed by two lavages with 1.2 ml of ethanol (96 to 100%) and an incubation of the tissue at 37°C to evaporate the remains of the ethanol. Volumes of 2 and 4 μl of DNA extracted from each sample were used for each PCR assay.
TABLE 2.
Species distribution and sensitivity of the new panfungal qPCR assay for 60 clinical samples included in the validation assaya
| Clinical group and fungal organism (no. of samples) | No. of organisms identified to species or genus level by: |
||
|---|---|---|---|
| Panfungal qPCRb | Molecular beaconc | Melting curved | |
| Proven IFI (37) | 29 | 18 (62.1%) | 15 (51.7%) |
| Aspergillus spp. (6) | 6 | 6 | 4 |
| Aspergillus fumigatus (2) | 2 | 2 | 1 |
| Aspergillus flavus (3) | 3 | 3 | 2 |
| Aspergillus penicilloides (1) | 1 | 1 | 1 |
| Mucormycetes (10) | 7 | 6 | 5 |
| Rhizopus oryzae (7) | 5 | 5 | 3 |
| Rhizopus microsporus (1) | 1 | 1 | 1 |
| Lichtheimia ramosa (1) | 1 | 0 | 1 |
| Rhizomucor pusillus (1) | 0 | 0 | 0 |
| Endemic mycoses (10) | 6 | 6 | 4 |
| Histoplasma capsulatum (8) | 4 | 4 | 2 |
| Coccidioides immitis (1) | 1 | 1 | 1 |
| Paracoccidioides brasiliensis (1) | 1 | 1 | 1 |
| Candida spp. (3) | 3 | 0 | 2 |
| Candida albicans (2) | 2 | 0 | 1 |
| Candida glabrata (1) | 1 | 0 | 1 |
| Other (8) | 7 | 0 | 0 |
| Fusarium sp. (1) | 1 | 0 | 0 |
| Cryptococcus neoformans (1) | 1 | 0 | 0 |
| Phoma exigua (1) | 0 | 0 | 0 |
| Bipolaris spicifera (1) | 1 | 0 | 0 |
| Microsphaeropsis arundinis (1) | 1 | 0 | 0 |
| Pythium insidiosum (1) | 1 | 0 | 0 |
| Cladophialophora bantiana (1) | 1 | 0 | 0 |
| Emmonsia crescens (1) | 1 | 0 | 0 |
| Probable IFI (23) | 21 | 10 (47.6%) | 7 (33.3%) |
| Aspergillus spp. (11) | 10 | 9 | 7 |
| Aspergillus fumigatus (6) | 6 | 6 | 4 |
| Aspergillus flavus (3) | 3 | 2 | 2 |
| Aspergillus terreus (1) | 1 | 1 | 1 |
| Aspergillus nomius (1) | 0 | 0 | 0 |
| Mucormycetes (1) | 1 | 1 | 0 |
| Rhizopus oryzae (1) | 1 | 1 | 0 |
| Candida spp. (6) | 6 | 0 | 0 |
| Candida albicans (2) | 2 | 0 | 0 |
| Candida parapsilosis (2) | 2 | 0 | 0 |
| Candida tropicalis (2) | 2 | 0 | 0 |
| Other (5) | 4 | 0 | 0 |
| Scedosporium sp. (1) | 1 | 0 | 0 |
| Aureobasidum pullulans (1) | 1 | 0 | 0 |
| Acremonium sp. (1) | 1 | 0 | 0 |
| Exophiala sp. (1) | 0 | 0 | 0 |
| Schizophyllum commune (1) | 1 | 0 | 0 |
| Total (60) | 50 | 28 (56%) | 22 (44%) |
Values in last three columns are numbers (percentage relative to positive samples by panfungal qPCR) of samples in which the organism was identified.
Overall identification of fungal species by the new panfungal qPCR assay by positive signal of Resolight dye followed by sequencing.
Identification of species or genus by positive signal of molecular beacon probes.
Identification of fungal species by melting curve analysis.
All PCR products obtained from clinical samples were run in 2% ethidium bromide-stained agarose gels (Sigma-Aldrich Química, Madrid, Spain) to verify amplification and the quality of the amplified products. Then, PCR products were purified by using the High Pure PCR product purification kit (Roche Diagnostics, Mannheim, Germany) and sequenced (ABI 3730 XL; Applied Biosystems, Madrid, Spain) with ITS1 and ITS2Deg2 primers to verify the results. The sequences obtained were compared with those available in the GenBank database (http://www.ncbi.nih.gov/GenBank/) and the sequence database belonging to the Mycology Department, National Centre for Microbiology, Instituto de Salud Carlos III, Madrid, Spain.
RESULTS
In vitro standardization of the qPCR assay.
The newly designed panfungal qPCR assay detected the DNAs from 11 fungal species included in the standardization assay. Each species was detected in the panfungal detection channel (FAM, 483 to 533 nm) and in the specific channel depending on the species tested (HEX, 523 to 568 nm, for Mucorales species; ROX, 558 to 610 nm, for Aspergillus spp.; or Cy5, 615 to 670 nm, for fungi causing endemic mycoses). Detection limits were established between 0.1 and 100 fg of DNA per μl, and CV values were within the acceptable limit of <5% in all cases with the exception of C. posadasii. Quantification was linear for all fungal species included in the assay, and the standard curve generated showed a coefficient of determination between 0.95 and 0.99 in all cases (Table 3).
TABLE 3.
Overview of standardization results of the 12 clinical strains belonging to 11 fungal species included in the new panfungal qPCR assaya
| Species | FAM (panfungal channel) |
HEX (Mucorales channel) |
ROX (Aspergillus sp. channel) |
Cy5 (endemic mycosis fungus channel) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S | R2 | CV (%) | DL (fg/μl) | S | R2 | CV (%) | DL (fg/μl) | S | R2 | CV (%) | DL (fg/μl) | S | R2 | CV (%) | DL (fg/μl) | |
| Rhizopus oryzae | −3.6 | 0.99 | 2.45 | 0.1 | −3.6 | 0.99 | 2.05 | 0.1 | ||||||||
| Rhizopus microsporus | −3.6 | 0.98 | 1.54 | 0.1 | −3.8 | 0.98 | 3.05 | 0.1 | ||||||||
| Mucor circinelloides | −3.4 | 0.98 | 3.76 | 0.1 | −3.7 | 0.98 | 4.67 | 1 | ||||||||
| Aspergillus fumigatus | −3.5 | 0.97 | 2.72 | 1 | −4.1 | 0.95 | 3.73 | 1 | ||||||||
| Aspergillus flavus | −3.8 | 0.98 | 3.69 | 1 | −4.1 | 0.98 | 3.05 | 1 | ||||||||
| Aspergillus terreus | −3.5 | 0.98 | 1.52 | 1 | −4.1 | 0.98 | 3.6 | 1 | ||||||||
| Histoplasma capsulatum var. capsulatum | −3.5 | 0.98 | 2.23 | 1 | −3.5 | 0.98 | 1.98 | 1 | ||||||||
| Histoplasma capsulatum var. duboisii | −3.4 | 0.98 | 2.21 | 10 | −3.3 | 0.98 | 2.16 | 10 | ||||||||
| Coccidioides immitis | −3.5 | 0.96 | 2.9 | 10 | −4.6 | 0.96 | 4.4 | 10 | ||||||||
| Coccidioides posadasii | −5.1 | 0.95 | 3.98 | 10 | −6.6 | 0.97 | 5.69 | 100 | ||||||||
| Blastomyces dermatitidis | −3.4 | 0.98 | 1.93 | 1 | −3.2 | 0.98 | 1.67 | 1 | ||||||||
| Paracoccidioides brasiliensis | −2.8 | 0.98 | 0.73 | 1 | −2.8 | 0.98 | 0.47 | 1 | ||||||||
S, slope of standard curve; R2, coefficient of determination; CV, coefficient of variation; DL, detection limit.
No cross-reactivity to DNA from other fungi or human or mouse was detected. Moreover, as expected, the MB-PanAspDeg probe and MB-PanMuc probes were available to detect species belonging to Aspergillus and Mucor genera, respectively.
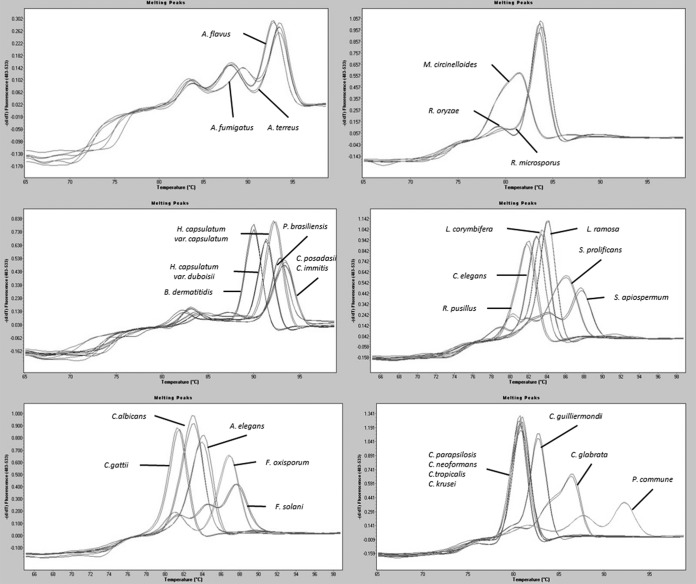
Melting curves were obtained for all species included in the standardization assay and checked visually for those with the best quality. All representative melting curves were collected as external melting standards, and the means of melting temperature values were noted (Table 4) with the aim to create a melting curve database, which was subsequently used to assess fungal species identification. In addition, melting curves of important fungal pathogens such as different species of Candida spp., Fusarium spp., and Scedosporium spp., which were not detected by the molecular beacon probes designed, were also included in the database (Fig. 1).
TABLE 4.
Melting temperature values obtained for each clinical fungal strain used in the standardization assay of the new panfungal qPCR assay
| Strain | Species |
Tm value (mean ± SD) |
||
|---|---|---|---|---|
| Tm1 | Tm2 | Tm3 | ||
| CNM-CM 2580 | A. fumigatus | 83.37 ± 0.13 | 89.18 ± 0.75 | 93.37 ± 0.38 |
| CNM-CM 3509 | A. flavus | 83.68 ± 0.69 | 88.06 ± 0.82 | 92.64 ± 0.08 |
| CNM-CM 3508 | A. terreus | 83.61 ± 0.63 | 87.89 ± 0.89 | 93.06 ± 0.24 |
| CNM-CM 3020 | R. oryzae | 78.73 ± 0.23 | 83.47 ± 0.19 | |
| CNM-CM 4244 | R. microsporus | 83.67 ± 0.22 | ||
| CNM-CM 2437 | M. circinelloides | 80.68 ± 0.43 | ||
| CNM-CM 2721 | H. capsulatum var. capsulatum | 92.57 ± 0.31 | ||
| CNM-CM 4626 | H. capsulatum var duboisii | 92.05 ± 1.41 | ||
| CNM-CM 7046 | C. immitis | 82.97 ± 0.70 | 92.89 ± 0.08 | |
| CNM-CM 2911 | C. posadasii | 82.37 ± 0.45 | 92.68 ± 0.47 | |
| CNM-CM 3114 | B. dermatitidis | 91.76 ± 0.81 | ||
| CNM-CM 2908 | P. brasiliensis | 90.85 ± 0.31 | ||
| CNM-CM 3169 | S. apiospermum | 83.68 ± 0.07 | 87.74 ± 0.06 | |
| CNM-CM 1627 | S. prolificans | 81.85 ± 0.06 | 85.84 ± 0.05 | |
| CNM-CM 3530 | F. solani | 81.30 ± 0.07 | 84.50 ± 0.1 | 87.66 ± 0.1 |
| CNM-CM 3197 | F. oxysporum | 81.05 ± 0.06 | 86.80 ± 0.08 | |
| CNM-CM 7046 | C. elegans | 82.84 ± 0.02 | ||
| CNM-CM 2751 | R. pusillus | 81.75 ± 0.13 | ||
| CNM-CM 7192 | P. commune | 81.75 ± 0.03 | 87.62 ± 0.01 | 92.21 ± 0.01 |
| CNM-CM 1722 | A. elegans | 83.81 ± 0.18 | ||
| CNM-CM 7130 | L. ramosa | 84.09 ± 0.01 | ||
| CNM-CM 7053 | L. corymbifera | 83.42 ± 0.06 | ||
| CNM-CL 8701 | C. albicans | 82.88 ± 0.03 | ||
| CNM-CL 5683 | C. parapsilosis | 80.68 ± 0.03 | ||
| CNM-CL 8796 | C. tropicalis | 80.79 ± 0.03 | ||
| CNM-CL 7523 | C. glabrata | 80.20 ± 0.05 | 86.53 ± 0.07 | |
| CNM-CL 7057 | C. krusei | 80.87 ± 0.01 | ||
| CNM-CL 7127 | C. guilliermondii | 82.75 ± 0.01 | ||
| CNM-CL 5801 | C. neoformans | 80.69 ± 0.01 | ||
| CNM-CL 5007 | C. gattii | 81.40 ± 0.03 | ||
FIG 1.
Melting curves for species included in the melting curve database constructed for species identification in the new panfungal qPCR assay.
Validation of the panfungal qPCR assay in clinical samples.
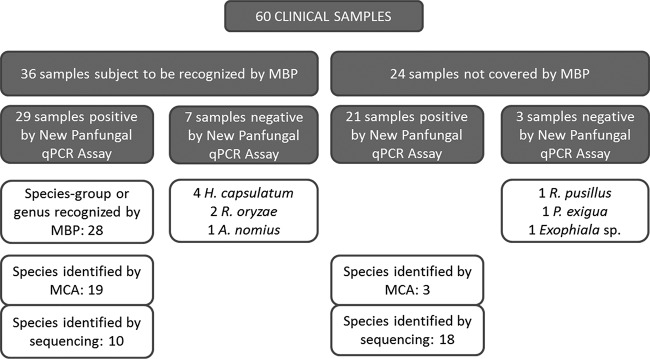
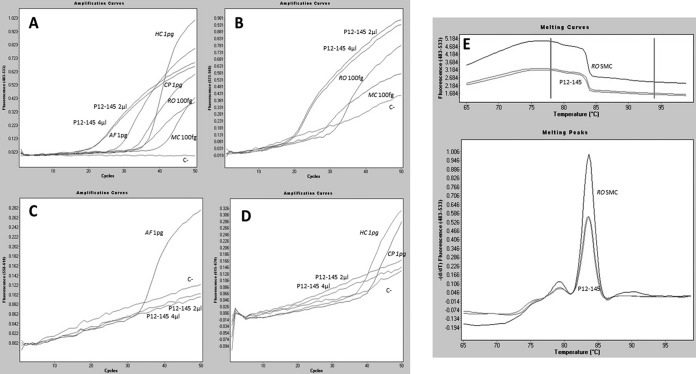
In order to assess the utility of this new panfungal qPCR assay, 60 clinical samples were tested. The assay was positive for 50 of the 60 clinical samples included in the validation assay (83.3%) with amplicon sequences confirming the species involved in the infection. Of the 36 cases in which causative species were susceptible to be recognized by any molecular beacon probe, we were able to identify the group of species involved in the IFI in 28 of them (77.8%). When the melting curve was analyzed in these cases, sequencing was avoided for 67.8% of them (19/28). Moreover, in the remaining 24 cases caused by species not covered by molecular probes, we were able to define the fungal species involved in the infection by melting curve analysis in two cases of infection by Candida spp. and one by Lichtheimia ramosa (Fig. 2). The assay detected fungal DNA in 29 of 37 (78.4%) samples of patients with proven IFI, 62.1% (18/29) of them with a positive signal of molecular beacon probes and 51.7% (15/29) with positive species identification achieved by melting curve analysis. In patients with probable IFI, the assay was positive for 20 of 23 of samples (87%) with 47.6% (10/23) of them detected by molecular beacon probes and 33.3% (7/23) reporting species identification by melting curve analysis. By species, the technique detected 94% of cases containing Aspergillus sp. DNA, 73% of cases caused by Mucorales species, 70% of cases in which endemic fungi caused the infection (7/10), 100% of cases harboring DNA from Candida spp. (9/9), and 85.5% of the cases containing DNA of rare and emerging species (11/13). Finally, 34 of the 44 biopsy specimens showed a positive result while all respiratory samples (8/8), all CSF samples (5/5), the aqueous humor fluid, and the nail sample gave a positive result. Results from the 60 clinical samples included in the study are detailed in Table 2. In Fig. 3, results for a positive sample are represented for each channel of detection of the real-time equipment.
FIG 2.
Chart summarizing the results of the validation assay of the new panfungal qPCR assay. Samples were differentiated in two blocks, those that could be recognized by probes and those that were not covered by any probe. Results for each group are detailed in white boxes. MCA, melting curve analysis; MBP, molecular beacon probe; P. exigua, Phoma exigua; A. nomius, Aspergillus nomius.
FIG 3.
Dependence of fluorescence signal on the number of cycles in the new panfungal qPCR assay for patient P12-145 with proven mucormycosis caused by R. oryzae. (A) Fluorescence detection in the FAM fluorescence channel, based on positive signal for the Resolight binding dye indicating panfungal amplification. (B) Fluorescence detection in the HEX fluorescence channel, showing specific detection of Rhizopus sp. and Mucor sp. DNA and 2 and 4 μl of DNA extracted from sample P12-145. (C) Fluorescence detection in the ROX fluorescence channel. The positive signal corresponded to the A. fumigatus positive control. (D) Fluorescence detection of Cy5 dye, showing H. capsulatum and C. posadasii positive-control amplification. (E) Melting peaks and melting curves detected in the FAM fluorescence channel obtained by melt curve genotyping analysis. R. oryzae standard melting curve matched with those melting curves obtained from the P12-145 clinical sample. RO, R. oryzae; MC, M. circinelloides; AF, A. fumigatus; HC, H. capsulatum; CP, C. posadasii; C-, negative control; SMC, standard melting curve.
DISCUSSION
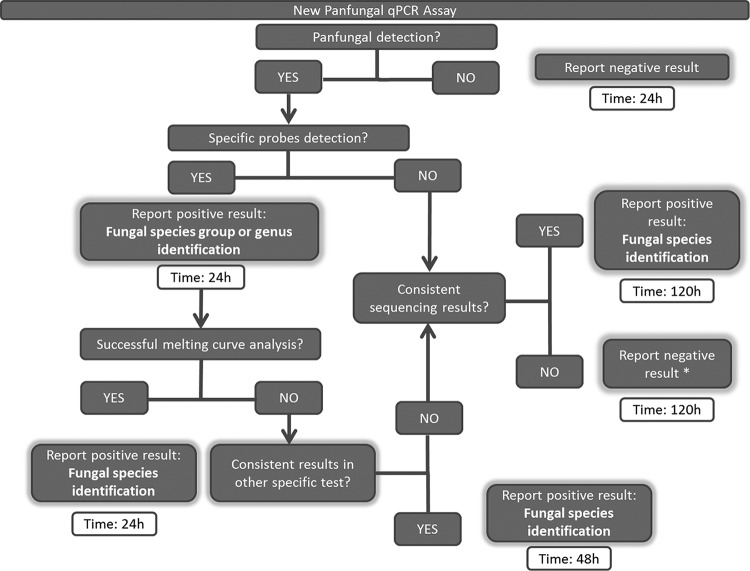
The aim of this work was to standardize and validate a new panfungal qPCR assay to improve diagnosis of IFIs when there is not a clear suspicion of the fungus involved in the disease. The usefulness of molecular methods based on PCR for the proper detection of fungal species in IFIs has been proven (25, 26). However, several limitations exist when the fungus causing the infection is unknown. In these cases, panfungal PCR assays have been used (27, 28), but they involve a delay in the response time because they require sequencing the amplicon. As recently described, in 30% of the samples studied from patients with IFI there was no evidence concerning the fungus implicated in the infection (24), highlighting the need for new methods. To overcome these limitations, several authors described the differentiation among fungal species by using melting curve analysis with panfungal primers (29, 30), demonstrating the capability of this technique to replace sequencing strategies, although sensitivity in these assays was limited. This new technique has been designed for the detection in one single tube of any fungal species and combines (i) panfungal detection by using a binding dye, (ii) specific detection by using molecular beacon probes, and (iii) species identification by using melting curve analysis. Merging all these techniques in the same assay eliminates the need for sequencing the amplicon and allows for an earlier identification of the species causing the disease. Figure 4 shows a diagram that explains the procedures to perform when using this new technique and also the time savings. Those samples with positive panfungal detection, positive probe detection, and a successful melting curve analysis may be reported as positive results within 24 h. When probe detection and melting curve analysis are negative, sequencing the amplicon becomes mandatory, increasing the response time to 120 h. However, this maximum time could be reduced to 48 h in those cases in which the species or genus was identified by molecular beacon probes by performing additional specific diagnostic tests. The selected design was directed to three groups of fungi (Aspergillus spp., Mucorales, and fungi causing endemic mycoses) by the use of five specific probes. Probes were designed using the sequence database belonging to the Mycology Department of the Instituto de Salud Carlos III, which contains a large set of ITS sequences representing a wide range of different fungal species. Other groups of fungi were not detected by using specific probes due to the limitations of the real-time PCR equipment. Despite this limitation, the technique is open and flexible to modifications, as new probes could be designed to focus on different groups of fungi depending on the needs of the laboratories, but it must be kept in mind that fluorescent channels available in qPCR equipment are limited.
FIG 4.
Schematic diagram of procedures to perform and time-to-result analysis for this new panfungal qPCR when there is no suspicion of species involved in the IFI. *, with the exception of those cases in which species or genus was detected by molecular beacon probes, for which samples should be retested or a new sample should be requested.
This new technique has been standardized with 12 clinical strains from 11 fungal species belonging to the collection of the Mycology Department of the Spanish Centre for Microbiology, Instituto de Salud Carlos III. During standardization, the sensitivity and reproducibility of this new panfungal qPCR were analyzed. Despite slight differences observed in detection limits described among different fungal species, the assay was efficient in detecting all fungal species targeted by the molecular beacon probes designed. The technique also exhibited high reproducibility and specificity. Moreover, other species of Mucor and Aspergillus genera, different from those used for the standardization process, were also detected by the MB-PanMuc and MB-AspDeg probes. Finally, a database with melting curves obtained from different fungal species was constructed. This database included all species used for the standardization process and also species belonging to important fungal pathogens such as Candida spp., Fusarium spp., and Scedosporium spp. that were not detected by specific probes but were included in the specificity test and collected as external melting standards.
The technique was validated by using 60 clinical samples from patients with proven and probable IFI. Samples were mainly biopsy specimens (44/60), followed by respiratory samples (8/60) and others (7/60). An overall sensitivity of 83.3% was achieved, showing similar sensitivity values in patients with proven and probable IFI. We were able to provide identification to the species level for 77.8% of those cases in which fungal species were covered by molecular beacon probes. In these cases, data from melting curves helped to define the species involved in 67.8% of the cases. Although melting curve analysis failed to give a definite diagnosis in nine of those “probe-positive” cases, the results provided important information that could help to perform additional specific PCR assays or to define an appropriate antifungal therapy for the patient. In addition, in three samples the species were identified by melting curve analysis because they were not targeted by molecular beacon probes. Finally, sequencing was mandatory for 56% of positive samples (28/50) mainly due to IFIs caused by species neither recognized by any probe nor included in the melting curve database constructed. Also, this technique showed some limitations, as certain species (i.e., C. parapsilosis, C. tropicalis, C. krusei, and C. neoformans) exhibited similar melting curves (Fig. 1) and had large standard deviations in Tm values (Table 4) that made them indistinguishable. In these cases, complementary techniques (specific PCRs, conventional tests, etc.) should be performed to assess fungal identification. Negative results were found only for biopsy samples; however, these results are biased, as these samples are overrepresented in comparison to respiratory and other samples included in the validation assay. Also, 5 of the 10 negative cases were found in paraffin-embedded biopsy specimens. Extraction of DNA suitable for amplification from these specimens is not optimized (31, 32). To discard the possibility that DNAs from those clinical samples with negative results were degraded, new DNA extraction was performed from five available clinical samples, but a positive result was obtained for only one sample. Of note, 7 of the negative samples harbored species that, theoretically, were susceptible to be detected by designed molecular beacon probes. However, species identification could not be achieved even by sequencing the amplicon. This could be due to a low fungal burden in the sample, the use of old, long-term-stored samples, or mixed infections, which have been described as main limitations of panfungal qPCR assays (3, 5).
In conclusion, this new panfungal qPCR assay is fast, sensitive, and specific for the detection of several fungal species in clinical samples, avoiding amplicon sequencing in many instances and shortening the time to definite diagnosis by up to 4 or 5 days. Further experiments are warranted in order to enlarge the melting curve database and expand the range of fungal species that can be detected by this technique.
ACKNOWLEDGMENT
We thank Frank Hodgkins for his careful reading and editing of the manuscript.
REFERENCES
- 1.Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, White TC. 2012. Hidden killers: human fungal infections. Sci Transl Med 4:165rv13. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 2.Cuenca-Estrella M, Bernal-Martinez L, Buitrago MJ, Castelli MV, Gomez-Lopez A, Zaragoza O, Rodriguez-Tudela JL. 2008. Update on the epidemiology and diagnosis of invasive fungal infection. Int J Antimicrob Agents 32(Suppl 2):S143–S147. doi: 10.1016/S0924-8579(08)70016-5. [DOI] [PubMed] [Google Scholar]
- 3.Buitrago MJ, Aguado JM, Ballen A, Bernal-Martinez L, Prieto M, Garcia-Reyne A, Garcia-Rodriguez J, Rodriguez-Tudela JL, Cuenca-Estrella M. 2013. Efficacy of DNA amplification in tissue biopsy samples to improve the detection of invasive fungal disease. Clin Microbiol Infect 19:E271–E277. doi: 10.1111/1469-0691.12110. [DOI] [PubMed] [Google Scholar]
- 4.Cuenca-Estrella M, Bassetti M, Lass-Florl C, Racil Z, Richardson M, Rogers TR. 2011. Detection and investigation of invasive mould disease. J Antimicrob Chemother 66(Suppl 1):i15–i24. doi: 10.1093/jac/dkq438. [DOI] [PubMed] [Google Scholar]
- 5.Khot PD, Fredricks DN. 2009. PCR-based diagnosis of human fungal infections. Expert Rev Anti Infect Ther 7:1201–1221. doi: 10.1586/eri.09.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kourkoumpetis TK, Fuchs BB, Coleman JJ, Desalermos A, Mylonakis E. 2012. Polymerase chain reaction-based assays for the diagnosis of invasive fungal infections. Clin Infect Dis 54:1322–1331. doi: 10.1093/cid/cis132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernal-Martinez L, Buitrago MJ, Castelli MV, Rodriguez-Tudela JL, Cuenca-Estrella M. 2013. Development of a single tube multiplex real-time PCR to detect the most clinically relevant Mucormycetes species. Clin Microbiol Infect 19:E1–E7. doi: 10.1111/j.1469-0691.2012.03976.x. [DOI] [PubMed] [Google Scholar]
- 8.Lu Q, Gerrits van den Ende AH, Bakkers JM, Sun J, Lackner M, Najafzadeh MJ, Melchers WJ, Li R, de Hoog GS. 2011. Identification of Pseudallescheria and Scedosporium species by three molecular methods. J Clin Microbiol 49:960–967. doi: 10.1128/JCM.01813-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gago S, Esteban C, Valero C, Zaragoza O, Puig de la Bellacasa J, Buitrago MJ. 2014. A multiplex real-time PCR assay for identification of Pneumocystis jirovecii, Histoplasma capsulatum, and Cryptococcus neoformans/Cryptococcus gattii in samples from AIDS patients with opportunistic pneumonia. J Clin Microbiol 52:1168–1176. doi: 10.1128/JCM.02895-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muraosa Y, Schreiber AZ, Trabasso P, Matsuzawa T, Taguchi H, Moretti ML, Mikami Y, Kamei K. 2014. Development of cycling probe-based real-time PCR system to detect Fusarium species and Fusarium solani species complex (FSSC). Int J Med Microbiol 304:505–511. doi: 10.1016/j.ijmm.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 11.Lau A, Chen S, Sorrell T, Carter D, Malik R, Martin P, Halliday C. 2007. Development and clinical application of a panfungal PCR assay to detect and identify fungal DNA in tissue specimens. J Clin Microbiol 45:380–385. doi: 10.1128/JCM.01862-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Babouee B, Goldenberger D, Elzi L, Lardinois D, Sadowski-Cron C, Bubendorf L, Savic PS, Battegay M, Frei R, Weisser M. 2013. Prospective study of a panfungal PCR assay followed by sequencing, for the detection of fungal DNA in normally sterile specimens in a clinical setting: a complementary tool in the diagnosis of invasive fungal disease? Clin Microbiol Infect 19:E354–E357. doi: 10.1111/1469-0691.12231. [DOI] [PubMed] [Google Scholar]
- 13.Trubiano JA, Dennison AM, Morrissey CO, Chua KY, Halliday CL, Chen SC, Spelman D. 2016. Clinical utility of panfungal polymerase chain reaction for the diagnosis of invasive fungal disease: a single center experience. Med Mycol 54:138–146. doi: 10.1093/mmy/myv092. [DOI] [PubMed] [Google Scholar]
- 14.Jordanides NE, Allan EK, McLintock LA, Copland M, Devaney M, Stewart K, Parker AN, Johnson PR, Holyoake TL, Jones BL. 2005. A prospective study of real-time panfungal PCR for the early diagnosis of invasive fungal infection in haemato-oncology patients. Bone Marrow Transplant 35:389–395. doi: 10.1038/sj.bmt.1704768. [DOI] [PubMed] [Google Scholar]
- 15.Landlinger C, Preuner S, Baskova L, van Grotel M, Hartwig NG, Dworzak M, Mann G, Attarbaschi A, Kager L, Peters C, Matthes-Martin S, Lawitschka A, van den Heuvel-Eibrink MM, Lion T. 2010. Diagnosis of invasive fungal infections by a real-time panfungal PCR assay in immunocompromised pediatric patients. Leukemia 24:2032–2038. doi: 10.1038/leu.2010.209. [DOI] [PubMed] [Google Scholar]
- 16.Arancia S, Sandini S, De Bernardis F, Fortini D. 2011. Rapid, simple, and low-cost identification of Candida species using high-resolution melting analysis. Diagn Microbiol Infect Dis 69:283–285. doi: 10.1016/j.diagmicrobio.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 17.Gago S, Zaragoza O, Cuesta I, Rodriguez-Tudela JL, Cuenca-Estrella M, Buitrago MJ. 2011. High-resolution melting analysis for identification of the Cryptococcus neoformans-Cryptococcus gattii complex. J Clin Microbiol 49:3663–3666. doi: 10.1128/JCM.01091-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ninghui G, Bing W, Wei R, Mengmeng L, Meiling C, Dongya M, Liqiong Y, Wencheng X. 2015. Application of PCR and high-resolution melting for rapid identification of yeasts routinely isolated in a clinical microbiology laboratory. Ann Clin Lab Sci 45:680–685. [PubMed] [Google Scholar]
- 19.Didehdar M, Khansarinejad B, Amirrajab N, Shokohi T. 2016. Development of a high-resolution melting analysis assay for rapid and high-throughput identification of clinically important dermatophyte species. Mycoses 59:442–449. doi: 10.1111/myc.12492. [DOI] [PubMed] [Google Scholar]
- 20.Lengerova M, Racil Z, Hrncirova K, Kocmanova I, Volfova P, Ricna D, Bejdak P, Moulis M, Pavlovsky Z, Weinbergerova B, Toskova M, Mayer J. 2014. Rapid detection and identification of mucormycetes in bronchoalveolar lavage samples from immunocompromised patients with pulmonary infiltrates by use of high-resolution melt analysis. J Clin Microbiol 52:2824–2828. doi: 10.1128/JCM.00637-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alonso M, Escribano P, Guinea J, Recio S, Simon A, Pelaez T, Bouza E, Garcia de Viedma D. 2012. Rapid detection and identification of Aspergillus from lower respiratory tract specimens by use of a combined probe-high-resolution melting analysis. J Clin Microbiol 50:3238–3243. doi: 10.1128/JCM.00176-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.White TJ, Bruns S, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal genes for phylogenetics, p 315–324. In Innis MA, Gelfand J, Sninsky J, White TJ (ed), PCR protocols: a guide to methods and applications. Academic Press, Inc., San Diego, CA. [Google Scholar]
- 23.De Pauw B, Walsh TJ, Donnelly JP, Stevens DA, Edwards JE, Calandra T, Pappas PG, Maertens J, Lortholary O, Kauffman CA, Denning DW, Patterson TF, Maschmeyer G, Bille J, Dismukes WE, Herbrecht R, Hope WW, Kibbler CC, Kullberg BJ, Marr KA, Munoz P, Odds FC, Perfect JR, Restrepo A, Ruhnke M, Segal BH, Sobel JD, Sorrell TC, Viscoli C, Wingard JR, Zaoutis T, Bennett JE. 2008. Revised definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clin Infect Dis 46:1813–1821. doi: 10.1086/588660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buitrago MJ, Bernal-Martinez L, Castelli MV, Rodriguez-Tudela JL, Cuenca-Estrella M. 2014. Performance of panfungal- and specific-PCR-based procedures for etiological diagnosis of invasive fungal diseases on tissue biopsy specimens with proven infection: a 7-year retrospective analysis from a reference laboratory. J Clin Microbiol 52:1737–1740. doi: 10.1128/JCM.00328-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Badiee P, Kordbacheh P, Alborzi A, Malekhoseini S, Ramzi M, Mirhendi H, Mahmoodi M, Shakiba E. 2009. Study on invasive fungal infections in immunocompromised patients to present a suitable early diagnostic procedure. Int J Infect Dis 13:97–102. doi: 10.1016/j.ijid.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 26.Halliday CL, Kidd SE, Sorrell TC, Chen SC. 2015. Molecular diagnostic methods for invasive fungal disease: the horizon draws nearer? Pathology 47:257–269. doi: 10.1097/PAT.0000000000000234. [DOI] [PubMed] [Google Scholar]
- 27.Lass-Florl C, Mutschlechner W, Aigner M, Grif K, Marth C, Girschikofsky M, Grander W, Greil R, Russ G, Cerkl P, Eller M, Kropshofer G, Eschertzhuber S, Kathrein H, Schmid S, Beer R, Lorenz I, Theurl I, Nachbaur D. 2013. Utility of PCR in diagnosis of invasive fungal infections: real-life data from a multicenter study. J Clin Microbiol 51:863–868. doi: 10.1128/JCM.02965-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sugawara Y, Nakase K, Nakamura A, Ohishi K, Sugimoto Y, Fujieda A, Monma F, Suzuki K, Masuya M, Matsushima Y, Wada H, Nobori T, Katayama N. 2013. Clinical utility of a panfungal polymerase chain reaction assay for invasive fungal diseases in patients with haematologic disorders. Eur J Haematol 90:331–339. doi: 10.1111/ejh.12078. [DOI] [PubMed] [Google Scholar]
- 29.Somogyvari F, Horvath A, Serly J, Majoros H, Vagvolgyi C, Peto Z. 2012. Detection of invasive fungal pathogens by real-time PCR and high-resolution melting analysis. In Vivo 26:979–983. [PubMed] [Google Scholar]
- 30.Bezdicek M, Lengerova M, Ricna D, Weinbergerova B, Kocmanova I, Volfova P, Drgona L, Poczova M, Mayer J, Racil Z. 2016. Rapid detection of fungal pathogens in bronchoalveolar lavage samples using panfungal PCR combined with high resolution melting analysis. Med Mycol 54:714–24. doi: 10.1093/mmy/myw032. [DOI] [PubMed] [Google Scholar]
- 31.Munoz-Cadavid C, Rudd S, Zaki SR, Patel M, Moser SA, Brandt ME, Gomez BL. 2010. Improving molecular detection of fungal DNA in formalin-fixed paraffin-embedded tissues: comparison of five tissue DNA extraction methods using panfungal PCR. J Clin Microbiol 48:2147–2153. doi: 10.1128/JCM.00459-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kocjan BJ, Hosnjak L, Poljak M. 2015. Commercially available kits for manual and automatic extraction of Nucleic acids from formalin-fixed, paraffin-embedded (FFPE) tissues. Acta Dermatovenerol Alp Pannonica Adriat 24:47–53. doi: 10.15570/actaapa.2015.12. [DOI] [PubMed] [Google Scholar]