Fig. 1.
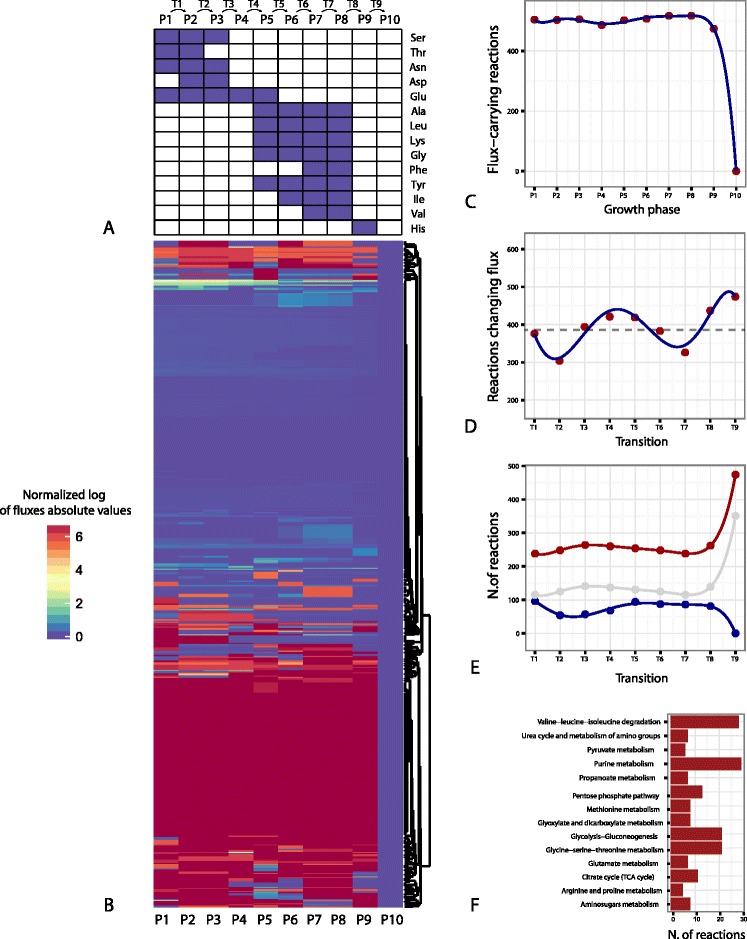
Summary of PhTAC125 genome-scale reprogramming following nutrients switching. a. The nutrients provided to the model in each different growth phase according to [14] b. Heat map with log values of fluxes across all the phases. c. Number of flux carrying reactions in each growth phase. d. Number of flux-changing reactions in each growth phase. The dashed line represents the average number of reactions carrying flux over all time points. e. Number of reactions whose flux is predicted to increase (blue line) and decrease (red line) following each shift in the nutrients provided; the black line accounts for those reactions whose flux is predicted to decrease not as an effect of an imposed reduced growth rate during simulations. f. functional annotation of reactions varying their flux across all the phases
