Fig. 6.
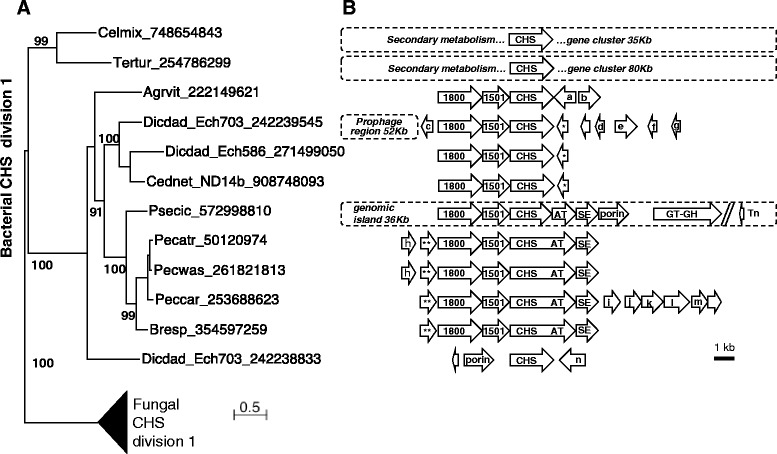
Phylogeny and genomic context of bacterial chitin synthases. a Phylogenetic relationships of bacterial chitin synthases. The ML tree is shown with numbers above the branches, indicating support in bootstrap analyses (100 replicates). The tree was rooted with division 1 fungal CHS from A. fumigatus and U. maydis. The Bayesian phylogenetic approach gave the exact same topology (Additional file 20: Figure S10). b Gene organization of each variable region containing chitin synthase. The limits of regions were obtained by comparison with ortholog regions in proximal species without the chs gene. Each represented region is aligned with the corresponding CHS in the phylogenetic tree. CHS in arrows is the chitin synthase domain and AT is the aminotransferase domain. 1800 and 1501: proteins with a domain of unknown function, DUF1800 (PF08811) and DUF1501 (PF07394) respectively; SE: sugar epimerase; a: transcriptional regulator with a sugar-binding domain; b: 2,4-dihydroxyhept-2-ene-1,7-dioic acid aldolase; c: OsmC family protein; d: DUF465; e: lipid A biosynthesis lauroyl (or palmitoleoyl) acyltransferase; f: cold-shock protein; g: DUF2511; h: efflux protein; i: gluconate-5-dehydrogenase; j: taurine dioxygenase; k: sulfotransferase NodH; l: aminotransferase; m: hydroxylase; and n: AraC family transcriptional regulator. The dotted rectangles correspond to regions detected as a prophage, a secondary metabolism cluster or a genomic island. Empty arrows and those with stars represent hypothetical proteins. Among them, groups of arrows with the same number of stars are orthologs
