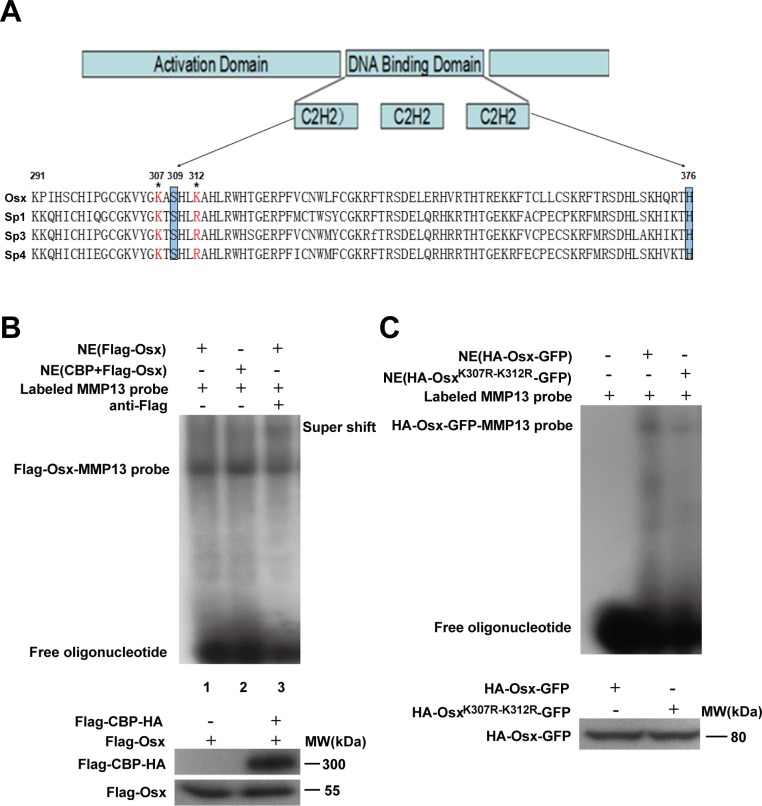
Figure 6. Acetylation increases the DNA binding activity of Osx.
A. Evolutionary conservation analysis of the acetylation sites in Osx. Schematic representation of Osx, its functional domains, the two acetylated lysine residues that were mutated to arginines and a partial polypeptide sequence alignment of Osx with Sp1, Sp3 and Sp4. The highlighted boxes indicate the zinc-finger domain (from amino acid 309 to 376). The acetylated lysines are indicated by asterisks. B. Nuclear extracts (NE) from HEK 293T cells transfected with Flag-Osx expression plasmid alone or together with Flag-CBP-HA were incubated with a 32P-labeled Osx-binding oligonucleotide (MMP-13 probe) in the absence (lane 1 and 2) or presence of anti-Flag antibodies (lane 3). The reaction mixtures were analyzed by EMSA. Osx and CBP proteins were detected by western blotting with an anti-Flag antibody. C. Nuclear extracts from HEK 293T cells transfected with HA-Osx-GFP or HA-OsxK307R-K312R-GFP expression plasmids were incubated in reaction mixtures containing 32P-labeled MMP-13 probe, resolved by electrophoreses and visualized by autoradiography. Osx and OsxK307R-K312R proteins were detected by western blotting with an anti-HA antibody. Experiments were repeated at least three times.