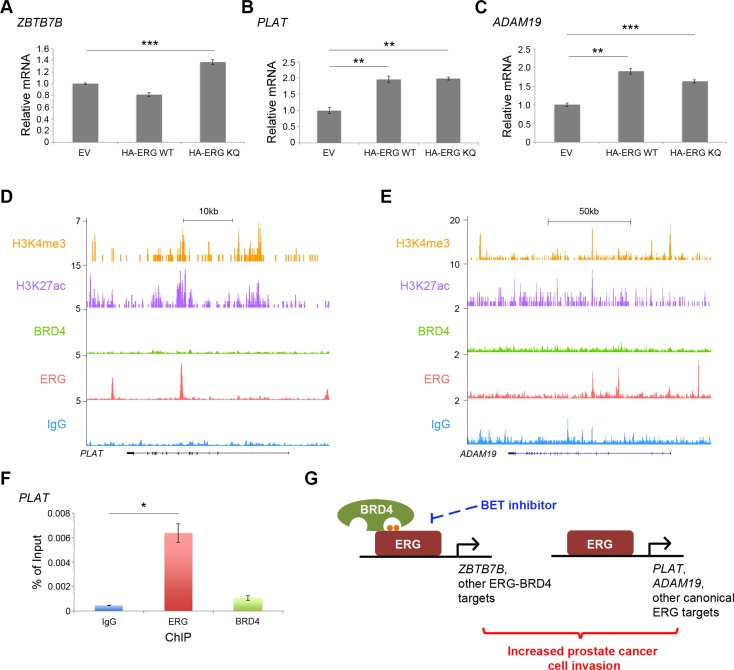
Figure 6. ERG-BRD4 co-targets and canonical ERG targets cooperate in prostatic cells.
(A) RT-qPCR for ZBTB7B in BPH-1 cells with over-expressed empty vector (EV), HA-ERG full-length wild-type (WT) or HA-ERG acetylation-mimic mutant (KQ). ***P < 0.001. (B) RT-qPCR for PLAT in BPH-1 cells with over-expressed EV, HA-ERG WT, or HA-ERG KQ. **P < 0.01. (C) RT-qPCR for ADAM19 in BPH-1 cells with over-expressed EV, HA-ERG WT, or HA-ERG KQ. **P < 0.01, ***P < 0.001. (D and E) Published BRD4 and ERG ChIP-seq data from VCaP cells [11]. Published H3K4me3 and H3K27ac ChIP-seq data from LNCaP cells [26] provide evidence indicating the regions of active transcription. (F) ERG and BRD4 ChIP-qPCR show the binding of ERG, but not BRD4 at the PLAT locus. *P < 0.05. (G) A hypothetical model. Acetylation-dependent interaction between ERG and BRD4 promotes transcription of co-target genes, and may contribute to increased cell invasion in combination with transcription of canonical ERG target genes. This ERG-BRD4 interaction can be targeted with BET inhibitors. Small orange dots represent acetylation sites on ERG protein.