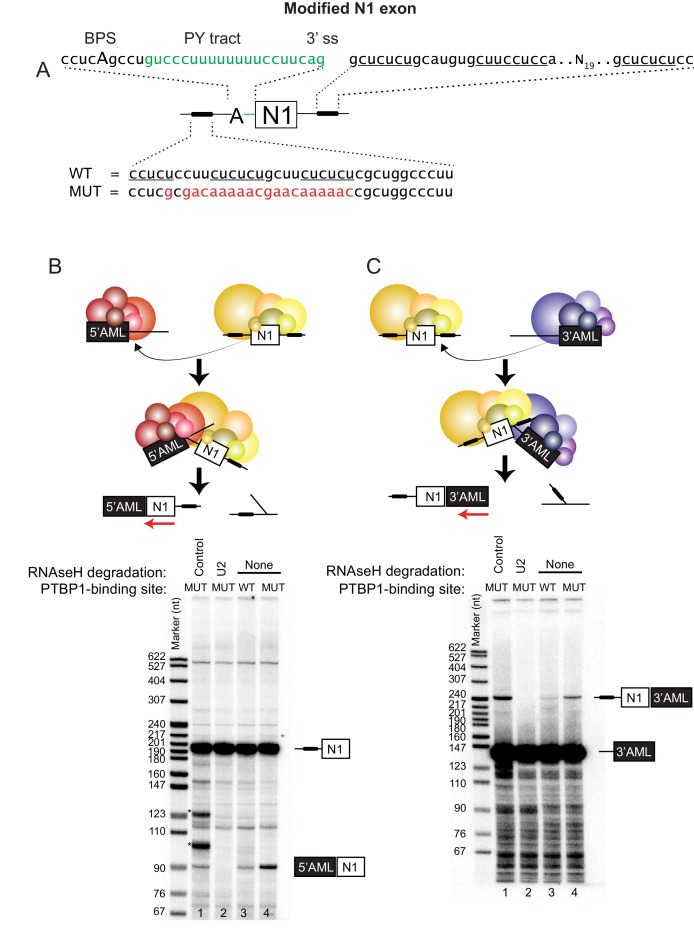
Figure 1. Distal PTBP1 binding flanking an alternative exon represses both splice sites.
(A) Diagram of the modified Src N1 exon RNA used in this study. PTBP1-binding sites are indicated as black bars. The upstream site (shown below) originally located within the polypyrimidine tract of the 3’ splice site was shifted to a position 25-nt upstream of the branch point sequence (BPS [Black, 1991]; cucAg). The polypyrimidine tract (PY tract) and 3’ splice site (3’ ss) of the Adenovirus Major Late (AML) first intron (highlighted in green) were added in place of the original N1 splice site. The downstream PTBP1-binding sites were left unchanged (shown above to the right). A mutation (MUT) of the upstream PTBP1-binding sites known to prevent PTBP1 binding is highlighted in red (Sharma et al., 2008; Amir-Ahmady et al., 2005). Nucleotides known to be important for PTBP1 binding are underlined. (B) Top panel: Diagram of the trans-splicing assay between the N1 exon and the 5’ splice site of the first AML intron (5’AML). Complexes assembled on either the WT or MUT N1 exon were mixed with the 5’AML complex and further incubated. Total RNA was extracted and assayed by primer extension analysis with a 32P labeled 3’ exon primer (red arrow). Bottom panel: primer extension products were resolved on 8% urea-PAGE. Spliced (90 nts.) and unspliced (195 nts) products are indicated. Bands marked with a black asterisk (*) resulted from an unexpected RNAse H cleavage of the N1 RNA by the control oligo, while a red asterisk (*) indicates self trans-splicing of the N1 exons. (C) Top panel: Diagram of the trans-splicing assay between the N1 exon and the downstream 3’ splice site of the first AML intron (3’AML). Reaction conditions were as in (B). Bottom panel: urea-PAGE of the primer extension assay as in (B), except using an oligo complementary to the 3’AML exon. Spliced (246 nts.) and unspliced (140 nts.) products are indicated.