FIGURE 3.
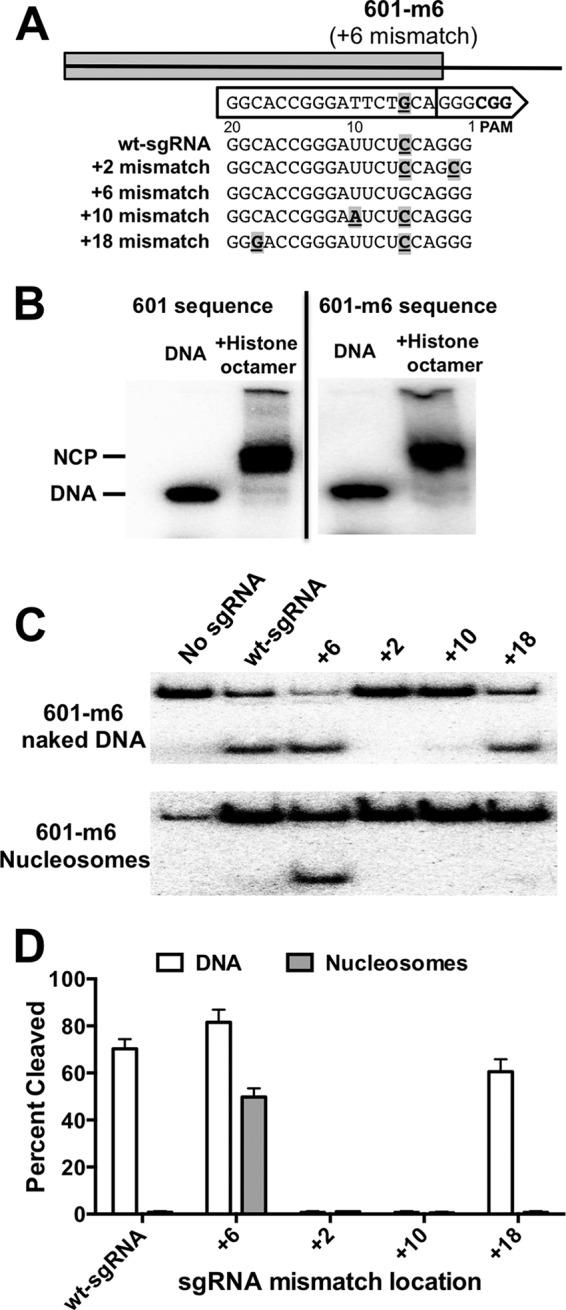
Cas9 activity with a mismatch sgRNA is restored with complementary mutation in 601 nucleosome substrate. A, diagram showing mismatch sgRNAs and complementary mutation in the 601 nucleosome substrate at position +6 nucleotides from the PAM site. B, representative reconstitutions of the wild-type 601 sequence and mutant DNA substrate (601-m6, with sequence change to complement the sgRNA +6 mismatch) into mononucleosomes analyzed by native polyacrylamide gel electrophoresis. NCP indicates the band corresponding to reconstituted nucleosome core particles. C, representative polyacrylamide gels showing cleavage of the 601-m6 naked DNA (top panel) and nucleosome (bottom panel) substrates after 30 min by Cas9 targeted by different sgRNAs used in the study. No sgRNA represents control samples with only the Cas9 enzyme (1 pmol) without the addition of an sgRNA. D, graph showing the percentage of cleavage of the 601-m6 DNA and nucleosome substrates by Cas9. Note that the +2, +10, and +18 sgRNAs have two mismatches with the 601-m6 substrate, because all have the equivalent of an additional +6 mismatch. Data points represent the average of at least three independent experiments, and error bars represent standard deviations.
