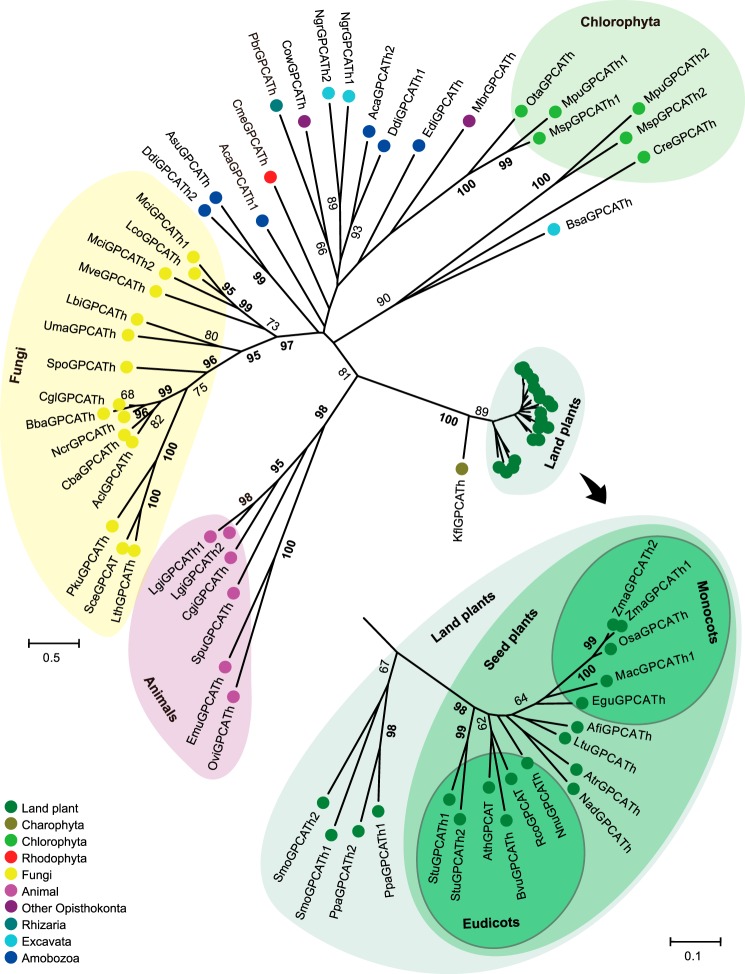
FIGURE 3.
Evolution of the GPCAT family in eukaryotes. GPCAT homologues from selected species across the eukaryotic clade were aligned and a radial unrooted tree built by maximum likelihood analysis using an LG+G model. The tree is drawn to scale, and branch lengths are measured in substitutions per site. Bootstrap values above 60 are shown (in percentages). Values of at least 95 are in boldface type. Inset, an expanded subtree for the land plant clade is shown. Aca, Acanthamoeba castellanii str. Neff; Acl, Aspergillus clavatus NRRL 1; Afi, Aristolochia fimbriata; Asu, Acytostelium subglobosum LB1; Ath, Arabidopsis thaliana; Atr, Amborella trichopoda; Bba, Beauveria bassiana D1-5; Bsa, Bodo saltans; Bvu, Beta vulgaris subsp. Vulgaris; Cba, Cladophialophora bantiana CBS 173.52; Cgi, Crassostrea gigas; Cgl, Chaetomium globosum CBS 148.51; Cme, Cyanidioschyzon merolae strain 10D; Cow, Capsaspora owczarzaki ATCC 30864; Cre, Chlamydomonas reinhardtii; Ddi, Dictyostelium discoideum AX4; Edi, Entamoeba dispar SAW760; Egu, Elaeis guineensis; Emu, Echinococcus multilocularis; Kfl, Klebsormidium flaccidum; Lbi, Laccaria bicolor S238N-H82; Lco, Lichtheimia corymbifera JMRC:FSU:9682; Lgi, Lottia gigantea; Lth, Lachancea thermotolerans CBS 6340; Ltu, Liriodendron tulipifera; Mac, Musa acuminata subsp. Malaccensis; Mbr, Monosiga brevicollis MX1; Mci, Mucorcircinelloides f. circinelloides 1006PhL; Mpu, Micromonas pusilla CCMP1545; Msp, Micromonas sp. RCC299; Mve, Mortierella verticillata NRRL 6337; Nad, Nuphar advena; Ncr, Neurospora crassa OR74A; Ngr, Naegleria gruberi strain NEG-M; Nnu, Nelumbo nucifera; Osa, Oryza sativa Indica group; Ota, Ostreococcus tauri; Ovi, Opisthorchis viverrini; Pbr, Plasmodiophora brassicae; Pku, Pichia kudriavzevii; Ppa, Physcomitrella patens; Rco, Ricinus communis; Sce, Saccharomyces cerevisiae S288c; Smo, Selaginella moellendorffii; Spo, Schizosaccharomyces pombe 972h; Spu, Strongylocentrotus purpuratus; Stu, Solanum tuberosum; Uma, Ustilago maydis 521; Zma, Zea mays. See supplemental Fig. S1 for protein alignment and sequence IDs.