Abstract
Objective
Carbohydrate-response element-binding protein (ChREBP) is the major transcription factor conferring glucose-induced gene expression in pancreatic islets, liver and adipose tissue. Recently, a novel ChREBP isoform, ChREBP-β, was identified in adipose tissue and found to be also expressed in islets and involved in glucose-induced beta cell proliferation. However, the physiological function of this less abundant β-isoform in the islet, and in diabetes, is largely unknown. The aims of the present study, therefore, were to determine how diabetes affects ChREBP-β and elucidate its physiological role in pancreatic beta cells.
Methods
Non-obese diabetic and obese, diabetic ob/ob mice were used as models of T1D and T2D and human islets and the rat INS-1 beta cell line were exposed to low/high glucose and used for ChREBP isoform-specific gain-and-loss-of-function experiments. Changes in ChREBP-β and ChREBP-α were assessed by qRT-PCR, immunoblotting, promoter luciferase, and chromatin immunoprecipitation studies.
Results
Expression of the ChREBP-β isoform was highly induced in diabetes and by glucose, whereas ChREBP-α was downregulated. Interestingly, ChREBP-β gain-of-function experiments further revealed that it was ChREBP-β that downregulated ChREBP-α through a negative feedback loop. On the other hand, ChREBP-β knockdown led to unabated ChREBP-α activity and glucose-induced expression of target genes, suggesting that one of the physiological roles of this novel β-isoform is to help keep glucose-induced and ChREBP-α-mediated gene expression under control.
Conclusions
We have identified a previously unappreciated negative feedback loop by which glucose-induced ChREBP-β downregulates ChREBP-α-signaling providing new insight into the physiological role of islet ChREBP-β and into the regulation of glucose-induced gene expression.
Keywords: Carbohydrate response element binding protein, Pancreatic islet, Glucose-induced gene expression, Transcription, Diabetes
Graphical abstract
Highlights
-
•
ChREBP-β is increased whereas expression of ChREBP-α is decreased in diabetes.
-
•
ChREBP-β downregulates ChREBP-α via a negative feedback loop in islets.
-
•
ChREBP-β thereby limits excessive glucose-induced ChREBP-α-mediated gene expression.
1. Introduction
Carbohydrate-response element-binding protein (ChREBP), also known as MLX-interacting protein-like (MLXIPL), is the major transcription factor conferring glucose-induced expression of glycolytic, gluconeogenic, and lipogenic genes, e.g. liver-type pyruvate kinase (L-PK), glycerol-3-phosphate dehydrogenase (GPDH), acetyl-CoA carboxylase (ACC) [1], [2], [3], [4], [5], and thioredoxin-interacting protein (TXNIP) [6], [7], and thereby plays a key role in metabolism. Glucose-induced transcriptional activity of ChREBP is thought to be mediated primarily by increased translocation into the nucleus, which, in turn, has been suggested to be regulated by dephosphorylation and/or release from cytoplasmic proteins [8], [9], [10], [11], [12]. In the nucleus, ChREBP induces transcription by binding to a highly conserved carbohydrate-response element (ChoRE) consisting of an E-box repeat located in the promoters of its target genes [6], [8], [13], [14]. While ChREBP was originally identified in liver, it has also been shown to be expressed in pancreatic beta cells of human, rat, and mouse [13], [15]. In 2012, a novel ChREBP isoform, ChREBP-β, was identified in adipose tissue [4]. ChREBP-β mRNA is transcribed from a ChoRE-containing promoter located 17 kb upstream of the ChREBP-α (former ChREBP) transcriptional start site, in which exon 1b is spliced to exon 2, bypassing exon 1a, and retaining the remainder of the ChREBP-α exons [4]. This results in a shorter transcript that produces a 687-amino acid protein (as opposed to the 864-amino acids of ChREBP-α) that lacks both nuclear export and nuclear localization signals as well as a domain that inhibits ChREBP-α transcriptional activity in low glucose [4]. As a result, ChREBP-β is constitutively active and shows increased nuclear localization and transcriptional activity at low and high glucose [4]. Therefore, although ChREBP-β mRNA expression is much less abundant than ChREBP-α, it has been suggested to be more potent [4]. Consistent with the ChoRE found in the ChREBP-β promoter (but not in the promoter of ChREBP-α), expression of the β-isoform is regulated by glucose and by ChREBP-α [4]. Recently, ChREBP-β mRNA was found to be also expressed in pancreatic islets although again at much lower levels as compared to the α-isoform and to be involved in glucose-stimulated beta cells proliferation [16]. However, the physiological function of ChREBP-β especially in the islet and under diabetic conditions has remained largely unknown. The goal of the present studies, therefore, was to investigate the role of ChREBP-β in pancreatic islets and in the context of diabetes.
2. Materials and methods
2.1. Cell culture and islet isolation
INS-1 cells were grown in RPMI 1640 (Invitrogen) with 11.1 mM glucose, 10% FBS, 1% penicillin/streptomycin, 1 mM sodium pyruvate, 2 mM l-glutamine, 10 mM HEPES, and 0.05 mM 2-mercaptoethanol. Mouse islets from 9-week old male obese and diabetic C57BL/6J lepob/ob (ob/ob) or lean control mice (JAX) [17] and ∼20-week or ∼10-week old female non-obese diabetic NOD/ShiLtJ or diabetes-resistant NOR/LtJ control mice (JAX) were isolated by collagenase digestion as described previously [18]. Mice were characterized as diabetic based upon 2 blood glucose measurements of >250 mg/dL at least one week apart. All mouse studies were approved by the University of Alabama at Birmingham Animal Care and Use Committee. Human islets were obtained through the Integrated Islet Distribution Program (IIDP), and islets from the same donor were always used as control and at least 3 different islet preparations were used per experiment.
2.2. Plasmid construction and transient transfection assays
The mouse ChREBP-α promoter region (2500 bp upstream Exon 1a) and ChREBP-β promoter region [4] were subcloned into the pGL3 enhancer vector (Promega) to generate luciferase reporter plasmids. To generate the ChREBP-β expression plasmid, mouse ChREBP-β cDNA was cloned and inserted into the pcDNA3.1/V5-His TOPO vector (Invitrogen). All constructs were verified by sequencing.
To determine the effects of glucose on ChREBP-α and β promoter activity, INS-1 cells were grown in 12-well plates and transfected with ChREBP-α or ChREBP-β promoter luciferase plasmids (0.4 μg/well) together with pRL-TK control plasmid (20 ng/well; Promega) in 5 mM glucose medium using DharmaFECT Duo transfection reagent (1 μl/well; Dharmacon/Thermo Scientific). After 24 h transfection, cells were treated with 5 mM or 25 mM glucose for 24 h and harvested; luciferase activity was determined by Dual Luciferase assay kit (Promega) as described previously [19]. For ChREBP-β overexpression experiments in INS-1 cells or human islets, transfections were performed at 11.1 mM glucose as previously described using 2 μg pcDNA/ChREBP-β and DharmaFECT Duo transfection reagent (6 μl/well) [17].
2.3. Small interfering RNA gene silencing
Isoform-specific siRNAs targeting the rat ChREBP-β coding region were designed and synthesized by Thermo Scientific as described previously [16]. INS-1 cells were grown in 6-well plates and transfected with specific siChREBP-β oligonucleotides or scrambled oligonucleotide using DharmaFECT 1 transfection reagent [19]. The final concentration of oligonucleotides used was 25 nM. Cells were harvested 48 h after transfection.
2.4. Quantitative real-time RT-PCR
Total RNA was extracted using miRNeasy Mini kit (Qiagen) according to the manufacturer's instructions. For regular quantitative real-time PCR, 1 μg RNA was reverse-transcribed to cDNA using the first strand cDNA synthesis kit (Roche). Results were corrected for the 18S ribosomal subunit (Applied Biosystems) run as an internal standard. All qRT-PCRs were performed on a LightCycler 480 System (Roche). qRT-PCR primers are shown in Supplemental Table S1.
2.5. Cell fractionation, immunoblotting and chromatin immunoprecipitation (ChIP)
Whole cell and nuclear protein extracts from INS-1 cells were prepared as described previously [20], [21]. Protein concentrations were measured by Pierce BCA protein assay (Thermo Fisher Scientific), and equal amounts of protein (20–50 μg/lane) were loaded. Actin was run as a loading control for whole cell extracts and USF2 for nuclear protein extracts. The following antibodies were used: Rabbit anti-ChREBP IgG targeting the internal region of 401–700 amino acids (1:500, sc-33764, Santa Cruz), Mouse anti-Actin IgG (1:5000, ab3280, Abcam), Rabbit anti-USF2 IgG (1:5000, sc-862, Santa Cruz), anti-rabbit IgG-HRP (1:3000, sc-2004; Santa Cruz), and anti-mouse IgG-HRP (1:3000, sc-2005; Santa Cruz). Bands were visualized by ECL plus (Amersham GE health) and quantified by ImageQuant.
Chromatin immunoprecipitation (ChIP) assays were performed as detailed previously [6] using Goat anti-ChREBP IgG (sc-21189, Santa Cruz) and primers flanking the ChREBP binding sites in the TXNIP and L-PK promoters (Supplemental Table S1).
2.6. Statistical analysis
Student's t-tests were used to calculate the significance of a difference between two groups. For data sets of more than 2 groups we performed one-way ANOVA calculations.
3. Results
3.1. Islet ChREBP-β is increased, whereas ChREBP-α is decreased in diabetes
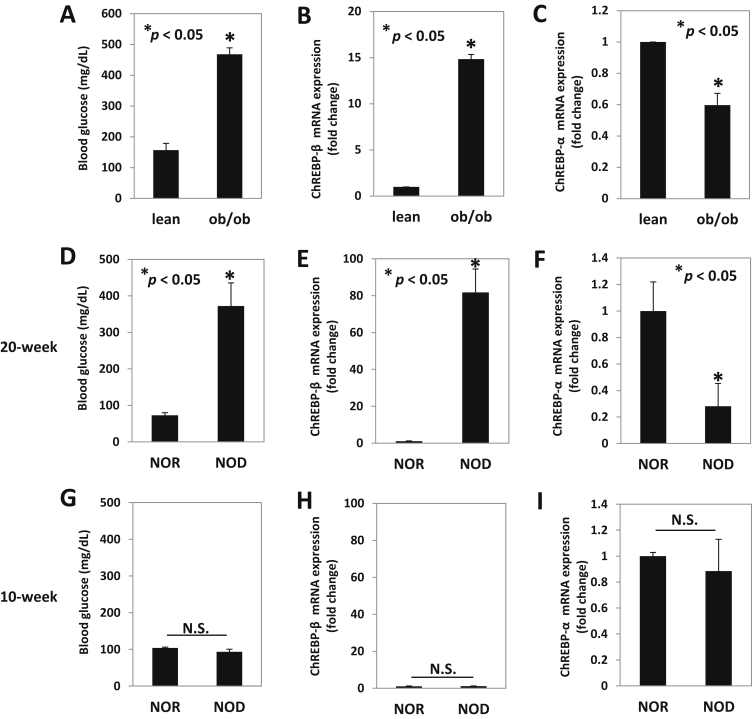
ChREBP-β was originally discovered in adipose tissue in 2012 [4], but we and others have found that this novel isoform of the ChREBP transcription factor is also expressed in pancreatic islets [16]. Therefore, we now investigated whether diabetes may affect islet ChREBP-β expression. Indeed, using obese, insulin-resistant, and diabetic ob/ob mice as a model of type 2 diabetes (Figure 1A), we found a dramatic 15-fold upregulation of ChREBP-β expression as compared to lean control mice (Figure 1B). Surprisingly, we also observed that the expression level of ChREBP-α was significantly lower in these ob/ob mice (Figure 1C). Interestingly, we saw very similar effects in non-obese diabetic NOD mice used as a model of type 1 diabetes (Figure 1D). Again, diabetes led to a striking induction of ChREBP-β (Figure 1E) but a downregulation of ChREBP-α (Figure 1F) in 20-week old female NOD mice as compared to non-diabetic control NOR mice. Of note, these effects were not seen in younger non-diabetic 10-week old female NOD mice, and no significant difference was observed in ChREBP-β or ChREBP-α expression between NOD and NOR islets (Figure 1G–I), indicating that the changes in the 20-week old animals were truly due to the diabetes. Also, since the main, common feature between ob/ob and NOD diabetes models is hyperglycemia, we hypothesized that the upregulation of ChREBP-β was due to the elevated glucose levels.
Figure 1.
Expression of ChREBP-β and ChREBP-α in islets of diabetic ob/ob and NOD mice. (A) Blood glucose, (B) ChREBP-β and (C) ChREBP-α expression was assessed using isoform-specific qRT-PCR in islets of diabetic and obese, ob/ob and non-diabetic, lean control mice. (D) Blood glucose, (E) ChREBP-β and (F) ChREBP-α expression was assessed in islets of 20-week old female non-obese diabetic NOD and diabetes-resistant NOR control mice as well as in islets of young female 10-week old non-diabetic NOD and NOR mice (G–I). Bars represent means ± SEM; n = 3–4 mice per group.
3.2. Glucose promotes islet ChREBP-β mRNA and protein levels but not ChREBP-α expression
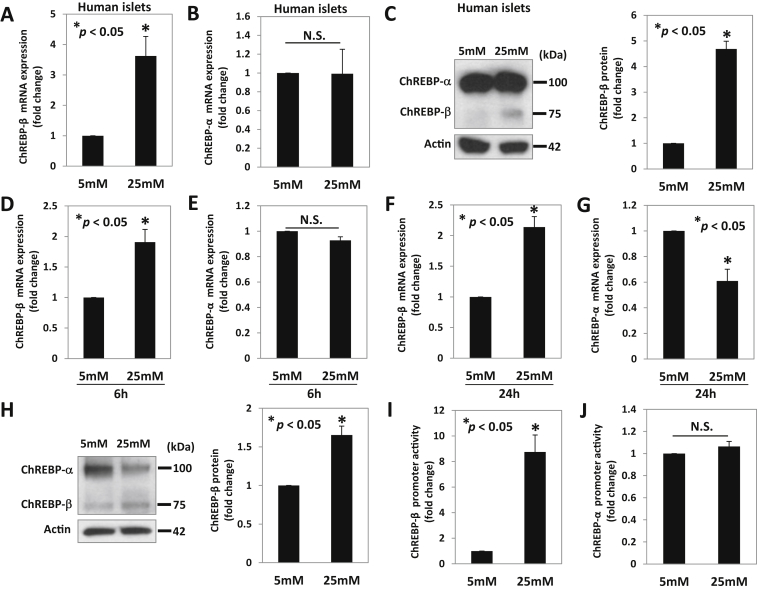
Indeed, high glucose was also able to upregulate ChREBP-β but not ChREBP-α expression in human islets (Figure 2A–B). So far, ChREBP-β has been studied primarily at the mRNA level as its expression level is much lower compared than that of ChREBP-α, and an isoform-specific antibody is not available [4] [16]. However, using an antibody targeting the internal region (401–700 amino acids) common to both ChREBP-α and ChREBP-β protein, we were able to detect two bands by immunoblotting, a larger and more prominent one corresponding to ChREBP-α (also confirmed by specific overexpression of this α-isoform) (Supplemental Figure S1A) and a smaller one, consistent with the predicted size of the 687-amino acid β-isoform, corresponding to ChREBP-β. This was confirmed by specific overexpression (Supplemental Figure S1B) and knockdown of ChREBP-β (Supplemental Figure S1C) as was the specificity of the ChREBP isoform-specific knockdown (Supplemental Figure. 2A–D). Using this approach, we next tested whether the observed changes in ChREBP-β expression in human islets were also translated into altered protein levels. Consistent with the mRNA results, we again found that glucose only increased ChREBP-β protein levels (Figure 2C). Similarly, using INS-1 cells, we found that at 6 h, glucose induced ChREBP-β mRNA expression, but ChREBP-α remained unchanged (Figure 2D–E). However, after 24 h incubation at high glucose, ChREBP-β mRNA and protein levels were increased, whereas ChREBP-α expression was decreased (Figure 2F–H) at this time point, indicating that the increase in ChREBP-β precedes the decrease of ChREBP-α. These results are consistent with our findings in the diabetic ob/ob and NOD mouse models. They are also in alignment with previous findings of increased ChREBP-β mRNA expression in response to glucose in adipose tissue and rat islets [4], [16] but now demonstrate that this increase also translates into significantly increased ChREBP-β protein levels.
Figure 2.
Effects of glucose on ChREBP-β and ChREBP-α mRNA and protein expression in human islets and INS-1 cells. ChREBP-β and α mRNA and protein expression was assessed in human islets incubated at 5 mM or 25 mM glucose for 24 h (A–C) or in INS-1 cells incubated at 5 mM or 25 mM glucose for 6 h (D–E) or 24 h (F–H) using qRT-PCR and immunoblotting. (I) ChREBP-β and (J) ChREBP-α promoter activity in response to glucose were assessed by luciferase assays in INS-1 cells transfected with ChREBP-α or β promoter luciferase plasmids together with pRL-TK control plasmid in 5 mM glucose medium and treated with 25 mM glucose for 24 h. Bars represent means ± SEM; at least 3 independent experiments were performed and representative immunoblots are shown.
To further assess whether glucose-induced differential regulation of the ChREBP isoforms is conferred at the transcriptional level, we also analyzed the promoter activities of ChREBP-β and ChREBP-α. Luciferase assays revealed that glucose dramatically induced ChREBP-β promoter activity (Figure 2I) but did not change the transcription of ChREBP-α (Figure 2J). These results are consistent with the previously reported transcriptional activation of ChREBP-β and the presence of a ChoRE in its promoter but not in the promoter of ChREBP-α [4], [16].
3.3. ChREBP-β downregulates ChREBP-α expression and transcriptional activity through a negative feedback loop
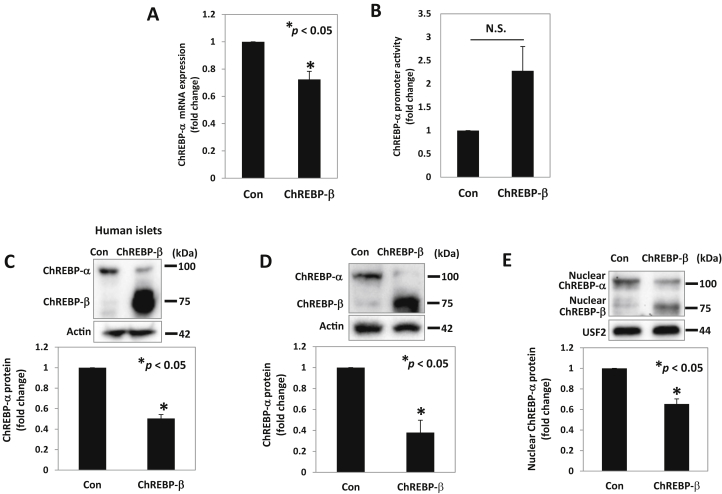
ChREBP-α has been shown to promote ChREBP-β expression [4], [16]. However, based on our findings that ChREBP-α was downregulated in islets of ob/ob and NOD mice as well as in response to glucose in INS-1 cells and that in each of these cases ChREBP-β was upregulated, we hypothesized that ChREBP-β, in turn, might also be regulating ChREBP-α. In fact, we found that ChREBP-β overexpression downregulated ChREBP-α mRNA expression (Figure 3A) but, in contrast, did not inhibit ChREBP-α promoter activity and even showed a trend towards increased transcriptional activity (Figure 3B). This makes it very unlikely that the observed ChREBP-β mediated ChREBP-α downregulation occurs at the transcriptional level. Importantly, we found that ChREBP-β overexpression also led to a significant downregulation of ChREBP-α protein levels in human islets (Figure 3C) and INS-1 cells (Figure 3D) and this translated into a significant decrease in transcriptionally active, nuclear ChREBP-α (Figure 3E), whereas no significant change was observed in cytosolic ChREBP-α (Supplemental Figure S3A), suggesting the existence of a negative feedback loop. While the exact molecular mechanism by which ChREBP-β downregulates ChREBP-α remains to be fully elucidated, the current results demonstrating decreased ChREBP-alpha mRNA levels as well as total and nuclear protein levels but no associated decrease in ChREBP-alpha promoter activity suggest that ChREBP-β may decrease ChREBP-α at a post-transcriptional level. To test the role of ChREBP-β in such a potential feedback loop we initiated a series of ChREBP-β knockdown experiments. Consistent with the ChREBP-β gain-of-function experiments (Figure 3D), we found that ChREBP-β loss-of-function resulted in the reverse, i.e. a small but significant increase in total ChREBP-α protein levels (Supplemental Figure S3B). However, the increase in ChREBP-α mRNA expression in response to ChREBP-β knockdown was only fully detectable under high glucose conditions (Supplemental Figure S3C), which is not surprising considering that baseline ChREBP-β expression is low and only increases in response to glucose.
Figure 3.
ChREBP-β effects on ChREBP-α expression and nuclear localization. INS-1 cells were maintained at 11.1 mM glucose medium, transfected with the ChREBP-β expression plasmid or control plasmid (Con) or cotransfected with the ChREBP-α promoter luciferase plasmid, and harvested 48 h after transfection. (A) ChREBP-α mRNA and (B) ChREBP-α promoter activity was assessed by qRT-PCR and luciferase assays, respectively. ChREBP-α protein levels were assessed by immunoblotting in (C) human islets or (D) INS-1 cells, 72 h or 48 h after transfection, respectively. (E) Nuclear ChREBP-α was assessed by immunoblotting after nuclear fractionation of INS-1 cells with or without ChREBP-β overexpression. Bars represent means ± SEM of at least 3 independent experiments and representative immunoblots are shown.
3.4. ChREBP-β knockdown leads to unopposed ChREBP-α activity and glucose-induced gene expression
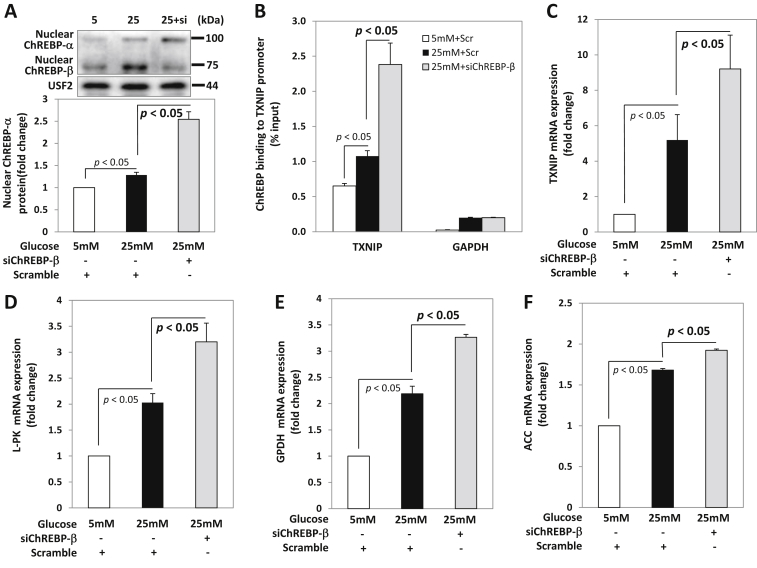
Together, these findings raised the possibility that, in addition to its well established ability to bind to ChoREs and thereby contribute to the transcription of glucose-induced genes [4] [16], ChREBP-β might help prevent excessive activation of the more abundant ChREBP-α isoform, particularly in response to acute glucose elevation. If ChREBP-β were truly conferring such a negative feedback loop in the context of high glucose, one would expect that knockdown of ChREBP-β would lead to disinhibition of ChREBP-α, increased nuclear localization, and enhanced glucose-induced expression of ChREBP target genes. Indeed, this is exactly what we observed. Incubation of INS-1 cells at high glucose led to a small but significant increase in nuclear ChREBP-α and a simultaneous increase in ChREBP-β, as expected. However, when this glucose-induced increase in ChREBP-β was prevented by transfection with specific siChREBP-β, a dramatic increase in nuclear ChREBP-α was observed as compared to cells transfected with scrambled control (Figure 4A). Moreover, ChREBP-β knockdown also led to a dramatic increase, far beyond what was seen with high glucose alone, in ChREBP-binding to the promoter of thioredoxin-interacting protein (TXNIP), a classical key glucose-induced ChREBP-α target gene (Figure 4B), whereas no binding was observed with IgG (Supplemental Fig. S3D), and this also resulted in a robust increase in TXNIP mRNA expression (Figure 4C). Furthermore, these effects were not restricted to TXNIP as very similar results were obtained with a number of other glucose-induced ChREBP-α target genes involved in glycolysis and fatty acid and lipid biosynthesis including L-PK (Figure 4D), GPDH (Figure 4E), as well as ACC (Figure 4F). This suggested that one of the physiological roles of this novel β-isoform of ChREBP is to help keep glucose-induced and ChREBP-α-mediated gene expression in check under high glucose conditions.
Figure 4.
Effects of ChREBP-β knockdown on ChREBP-α and glucose-induced target gene expression. INS-1 cells were transfected with specific siRNA oligonucleotides for rat ChREBP-β (siChREBP-β) or scrambled control (Scramble) for 48 h and then treated with 25 mM glucose for 12 h before being harvested. (A) Nuclear ChREBP-α protein levels, (B) ChREBP binding to the TXNIP promoter were assessed by immunoblotting and ChIP assays. mRNA expression of (C) TXNIP, (D) L-PK (E) GPDH, and (F) ACC was assessed by qRT-PCR. Bars represent means ± SEM of at least 3 independent experiments and a representative immunoblot is shown.
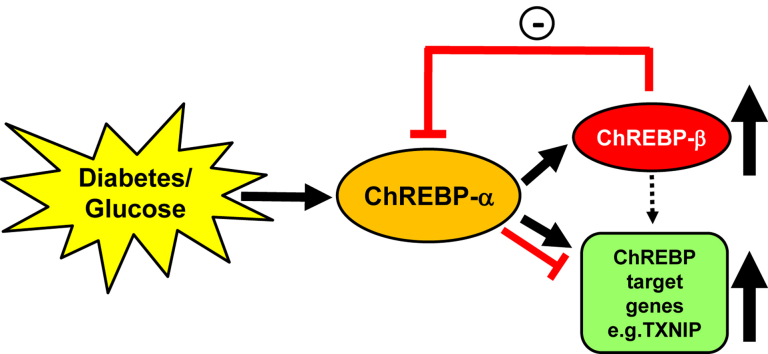
Taken together, these data support the novel notion that in diabetes and in response to high glucose, ChREBP-α is activated and enters the nucleus to mediate transcription of glucose-induced target genes; however, ChREBP-α also stimulates transcription of its less abundant β-isoform, ChREBP-β, and ChREBP-β, in turn, provides a negative feedback loop downregulating ChREBP-α and the transcription of ChREBP target genes such as TXNIP, L-PK, GPDH, and ACC, thereby limiting excessive glucose-induced gene expression (Graphical Abstract).
4. Discussion
The results of the present study reveal for the first time that the novel β-isoform of ChREBP is upregulated, whereas ChREBP-α is downregulated, in vivo in mouse models of type 1 and type 2 diabetes. Furthermore, the results demonstrate that glucose-induced ChREBP-β downregulates ChREBP-α expression and transcriptional activity, suggesting that ChREBP-β may provide a negative feedback loop that keeps glucose-induced, ChREBP-α-mediated gene expression under control.
ChREBP-β was originally identified in adipose tissue and found to be regulated at the transcriptional level by ChREBP-α, which binds to a ChoRE 17 kb upstream of the ChREBP-β transcriptional start site. This regulation of ChREBP-β by ChREBP-α has also been demonstrated very nicely in pancreatic islets [16]. Now, our data indicate that ChREBP-β, in turn, regulates ChREBP-α and thereby provides a negative feedback loop. Of note, this effect was observed in INS-1 cells as well as in human islets and ChREBP-α mRNA, total protein, and nuclear protein levels were reduced in response to ChREBP-β. This notion of ChREBP-β downregulating ChREBP-α is also in alignment with our observation that increased ChREBP-β expression in response to glucose was associated with decreased ChREBP-α levels and that ChREBP-β expression was significantly increased in islets of diabetic ob/ob and NOD mice while ChREBP-α was dramatically decreased as compared to non-diabetic controls. Consistent with the relatively low abundance of ChREBP-β in primary islets, total ChREBP, reflecting primarily changes in the predominant ChREBP-α isoform, was also significantly reduced in diabetic NOD islets (p < 0.05, data not shown). Intriguingly, total ChREBP gene expression was also reduced in laser-captured islets from human T1D organ donors as compared to non-diabetic donors (Ivan C. Gerling, personal communication), further supporting the pathophysiological significance of our observations.
Moreover, we found that knockdown of ChREBP-β under high glucose conditions and with that, inhibition of this negative feedback loop resulted in a dramatic increase in nuclear ChREBP-α and ChREBP-mediated transcription of classical target genes including TXNIP, L-PK, GPDH, and ACC. Of note, we have found previously that increased TXNIP expression has deleterious effects on beta cell biology, promotes pancreatic beta cell apoptosis and islet amyloid polypeptide expression, is a critical factor in glucotoxicity-induced beta cell death, and impairs insulin production and beta cell function [14], [17], [19], [20], [22]. Together, these results support the notion that glucose-induced ChREBP-β constitutes a compensatory cellular response to limit excessive ChREBP-α signaling and glucose-induced expression of potentially detrimental genes such as TXNIP.
The fact that a previous report has found decreased expression of ChREBP target gene such as TXNIP after 4 days of ChREBP-β knockdown in the context of high glucose [16] may be due to the difference in timing. In an acute or short-term setting as in our case, the contribution of ChREBP-β to overall ChREBP-mediated transcription of target genes is expected to be low given its relative low expression level compared to ChREBP-α. On the other hand, it is conceivable that after chronic or long-term induction by elevated glucose, the contribution of ChREBP-β to overall ChREBP-mediated gene transcription may increase. Such a scenario would be consistent with the notion that in the short-term, glucose-induced expression of ChREBP-β may provide a protective compensatory mechanism to keep glucose-induced gene expression under control, but in response to chronically elevated glucose levels, constitutively active ChREBP-β itself may start contributing to glucose-induced gene expression, including that of TXNIP, L-PK, and GPDH. This would also be in alignment with the fact that in diabetes, TXNIP has been shown to be elevated in pancreatic islets [6], [14], [20], [23]; yet, we now have discovered that ChREBP-α is downregulated. Overall though, the regulatory role of ChREBP-β identified in the present studies seems to be more in line with its relative low expression level, especially in primary islets, than with any major direct role as a transcription factor conferring glucose-induced gene expression under physiological conditions.
Overexpression of a constitutively active, truncated form of ChREBP-α has been used previously to elucidate the effects of ChREBP and overcome the problem of native ChREBP-α being excluded from the nucleus in the absence of high glucose. As such, these studies have greatly contributed to our understanding of the detrimental effects excessive ChREBP-α can have on beta cell biology [24]. In a recent report, overexpression of this constitutively active, truncated form of ChREBP-α failed to regulate endogenous ChREBP-α [25]. While this truncated form of ChREBP-α resembles ChREBP-β in that it lacks a N-terminal domain and, therefore, can freely enter the nucleus, it lacks amino acids 1–196, resulting in a 667-amino acid protein, whereas ChREBP-β only lacks amino acids 1–176 giving rise to a 687-amino acid protein. Therefore, it is not surprising that the two distinct proteins may behave differently, especially when it comes to effects that go beyond transcription.
5. Conclusion
We have found that the novel ChREBP-β isoform is increased in diabetic islets, whereas ChREBP-α is decreased. We have also identified a previously unappreciated negative feedback loop by which diabetes or glucose-induced ChREBP-β downregulates ChREBP-α and ChREBP-α-mediated transcription of important glucose-induced metabolic genes. These results suggest that ChREBP-β provides a compensatory response limiting excessive ChREBP-α-mediated gene expression in the context of diabetes or high glucose conditions and thereby shed new light on the physiological role of this novel β-isoform of ChREBP in pancreatic islets and on the regulation of glucose-induced gene expression in general.
Author contribution
G.J. performed and analyzed most of the experiments and wrote a manuscript draft, J.C. was responsible for the mouse studies and islet isolations, G.X. helped with some of the molecular biology, and A.S. conceived the work, supervised the studies, and revised the manuscript.
Acknowledgements
The work was supported by grants to A.S. from the National Institutes of Health (R01DK-078752, UC4DK104204) and JDRF (3-SRA-2014-302-M-R). These funding sources had no involvement in the study design, collection analysis, or interpretation of the data.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molmet.2016.09.010.
Conflict of interest
None declared.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Iizuka K., Bruick R.K., Liang G., Horton J.D., Uyeda K. Deficiency of carbohydrate response element-binding protein (ChREBP) reduces lipogenesis as well as glycolysis. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:7281–7286. doi: 10.1073/pnas.0401516101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ma L., Robinson L.N., Towle H.C. ChREBP*Mlx is the principal mediator of glucose-induced gene expression in the liver. The Journal of Biological Chemistry. 2006;281:28721–28730. doi: 10.1074/jbc.M601576200. [DOI] [PubMed] [Google Scholar]
- 3.Iizuka K., Wu W., Horikawa Y., Saito M., Takeda J. Feedback looping between ChREBP and PPARalpha in the regulation of lipid metabolism in brown adipose tissues. Endocrine Journal. 2013;60:1145–1153. doi: 10.1507/endocrj.ej13-0079. [DOI] [PubMed] [Google Scholar]
- 4.Herman M.A., Peroni O.D., Villoria J., Schön M.R., Abumrad N.A., Blüher M. A novel ChREBP isoform in adipose tissue regulates systemic glucose metabolism. Nature. 2012;484:333–338. doi: 10.1038/nature10986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kursawe R., Caprio S., Giannini C., Narayan D., Lin A., D'Adamo E. Decreased transcription of ChREBP-alpha/beta isoforms in abdominal subcutaneous adipose tissue of obese adolescents with prediabetes or early type 2 diabetes: associations with insulin resistance and hyperglycemia. Diabetes. 2013;62:837–844. doi: 10.2337/db12-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cha-Molstad H., Saxena G., Chen J., Shalev A. Glucose-stimulated expression of Txnip is mediated by carbohydrate response element-binding protein, p300, and histone H4 acetylation in pancreatic beta cells. The Journal of Biological Chemistry. 2009;284:16898–16905. doi: 10.1074/jbc.M109.010504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kibbe C., Chen J., Xu G., Jing G., Shalev A. FOXO1 competes with carbohydrate response element-binding protein (ChREBP) and inhibits thioredoxin-interacting protein (TXNIP) transcription in pancreatic beta cells. The Journal of Biological Chemistry. 2013;288:23194–23202. doi: 10.1074/jbc.M113.473082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rufo C., Teran-Garcia M., Nakamura M.T., Koo S.H., Towle H.C., Clarke S.D. Involvement of a unique carbohydrate-responsive factor in the glucose regulation of rat liver fatty-acid synthase gene transcription. The Journal of Biological Chemistry. 2001;276:21969–21975. doi: 10.1074/jbc.M100461200. [DOI] [PubMed] [Google Scholar]
- 9.Wang H., Wollheim C.B. ChREBP rather than USF2 regulates glucose stimulation of endogenous L-pyruvate kinase expression in insulin-secreting cells. The Journal of Biological Chemistry. 2002;277:32746–32752. doi: 10.1074/jbc.M201635200. [DOI] [PubMed] [Google Scholar]
- 10.Kawaguchi T., Takenoshita M., Kabashima T., Uyeda K. Glucose and cAMP regulate the L-type pyruvate kinase gene by phosphorylation/dephosphorylation of the carbohydrate response element binding protein. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:13710–13715. doi: 10.1073/pnas.231370798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Filhoulaud G., Guilmeau S., Dentin R., Girard J., Postic C. Novel insights into ChREBP regulation and function. Trends in Endocrinology and Metabolism. 2013;24:257–268. doi: 10.1016/j.tem.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 12.Noordeen N.A., Meur G., Rutter G.A., Leclerc I. Glucose-induced nuclear shuttling of ChREBP is mediated by sorcin and Ca(2+) ions in pancreatic beta-cells. Diabetes. 2012;61:574–585. doi: 10.2337/db10-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamashita H., Takenoshita M., Sakurai M., Bruick R.K., Henzel W.J., Shillinglaw W. A glucose-responsive transcription factor that regulates carbohydrate metabolism in the liver. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:9116–9121. doi: 10.1073/pnas.161284298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Minn A.H., Hafele C., Shalev A. Thioredoxin-interacting protein is stimulated by glucose through a carbohydrate response element and induces beta-cell apoptosis. Endocrinology. 2005;146:2397–2405. doi: 10.1210/en.2004-1378. [DOI] [PubMed] [Google Scholar]
- 15.Metukuri M.R., Zhang P., Basantani M.K., Chin C., Stamateris R.E., Alonso L.C. ChREBP mediates glucose-stimulated pancreatic beta-cell proliferation. Diabetes. 2012;61:2004–2015. doi: 10.2337/db11-0802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang P., Kumar A., Katz L.S., Li L., Paulynice M., Herman M.A. Induction of the ChREBPbeta isoform is essential for glucose-stimulated beta cell proliferation. Diabetes. 2015;64:4158–4170. doi: 10.2337/db15-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu G., Chen J., Jing G., Shalev A. Thioredoxin-interacting protein regulates insulin transcription through microRNA-204. Nature Medicine. 2013;19:1141–1146. doi: 10.1038/nm.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Minn A.H., Lan H., Rabaglia M.E., Harlan D.M., Peculis B.A., Attie A.D. Increased insulin translation from an insulin splice-variant overexpressed in diabetes, obesity, and insulin resistance. Molecular Endocrinology. 2005;19:794–803. doi: 10.1210/me.2004-0119. [DOI] [PubMed] [Google Scholar]
- 19.Jing G., Westwell-Roper C., Chen J., Xu G., Verchere C.B., Shalev A. Thioredoxin-interacting protein promotes islet amyloid polypeptide expression through miR-124a and FoxA2. The Journal of Biological Chemistry. 2014;289:11807–11815. doi: 10.1074/jbc.M113.525022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen J., Saxena G., Mungrue I.N., Lusis A.J., Shalev A. Thioredoxin-interacting protein: a critical link between glucose toxicity and beta-cell apoptosis. Diabetes. 2008;57:938–944. doi: 10.2337/db07-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saxena G., Chen J., Shalev A. Intracellular shuttling and mitochondrial function of thioredoxin-interacting protein. The Journal of Biological Chemistry. 2010;285:3997–4005. doi: 10.1074/jbc.M109.034421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shalev A. Minireview: thioredoxin-interacting protein: regulation and function in the pancreatic beta-cell. Molecular Endocrinology. 2014;28:1211–1220. doi: 10.1210/me.2014-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J., Hui S.T., Couto F.M., Mungrue I.N., Davis D.B., Attie A.D. Thioredoxin-interacting protein deficiency induces Akt/Bcl-xL signaling and pancreatic beta-cell mass and protects against diabetes. FASEB Journal. 2008;22:3581–3594. doi: 10.1096/fj.08-111690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poungvarin N., Lee J.K., Yechoor V.K., Li M.V., Assavapokee T., Suksaranjit P. Carbohydrate response element-binding protein (ChREBP) plays a pivotal role in beta cell glucotoxicity. Diabetologia. 2012;55:1783–1796. doi: 10.1007/s00125-012-2506-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sae-Lee C., Moolsuwan K., Chan L., Poungvarin N. ChREBP regulates itself and metabolic genes implicated in lipid accumulation in beta-cell line. PLoS One. 2016;11:e0147411. doi: 10.1371/journal.pone.0147411. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.