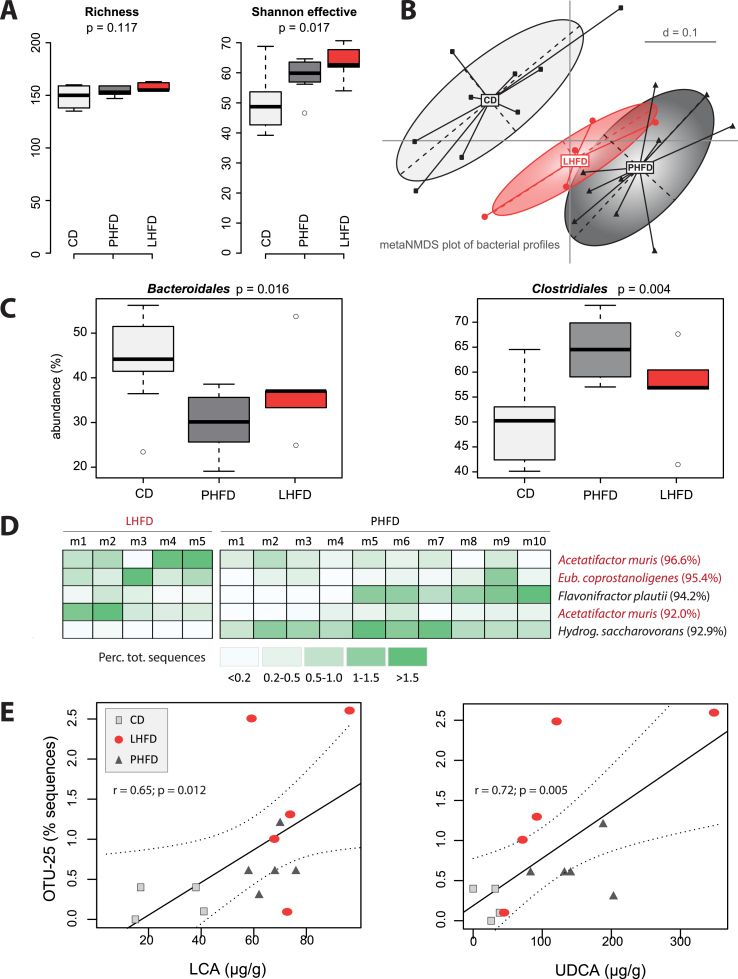
Figure 5.
Specific dominant gut bacteria are affected by dietary fat source. Metagenomic DNA isolated from fecal samples (n = 24) was used for amplification of the V3/V4 region of 16S rRNA genes and subsequent sequencing using the Illumina technology. Sequences were analyzed using in-house developed pipelines as described in detail in the methods section. (A) Alpha-diversity analysis. (B) Multidimensional scaling showing differences in diversity between samples (beta-diversity) based on general UniFrac distances. (C) Box plots showing relative sequence abundance of taxonomic groups that were significantly different between mice fed the CD or HFD. (D) Phylotype analysis shown as heatmap of OTU abundances which were significantly different between the two HFD. The identity of OTUs was obtained using EzTaxon based on sequences of approximately 380 bp [64]. Best hits are shown with corresponding sequences similarity. (E) Pearson correlation analysis of Acetatifactor sp. against cecal bile acid concentrations.