Fig. 7.
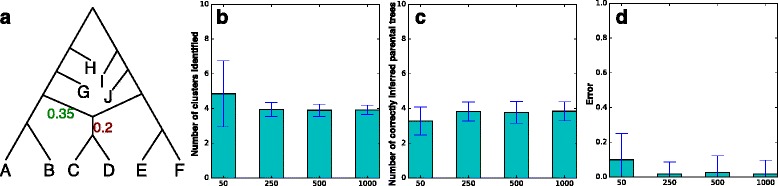
Performance of the clustering approach on the simulated data as a function of the number of gene trees. a The phylogenetic network used in the simulations. The lengths of the two reticulation edges were set to 0. The length of the edge going out of the reticulation node was set to 0.2. The inheritance probability of the left reticulation edge was set to 0.35. Ten networks were generated from this network by setting the length of each other internal branch to a random number uniformly sampled in the range [0.7,1.3]. b The number of clusters identified (averaged over 300 data sets for each bar). c The number of correctly inferred parental trees (out of the maximum of four parental trees). d The error between the set of inferred trees from the identified clusters and the set of four parental trees of the network. The x-axis in panels (b)-(d) corresponds to the number of gene trees
