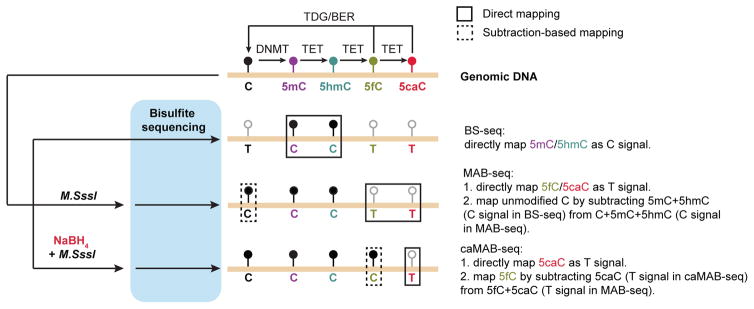
Figure 1. Schematic diagram of BS-seq, MAB-seq and caMAB-seq.
In mammalian cells, de novo and maintenance DNA methyltransferases (DNMTs) methylate unmodified cytosines (C) to generate 5-methylcytosines (5mC). Ten-eleven translocation (TET) family of DNA dioxygenase (TET1–3) is capable of iteratively oxidizing 5mC and its derivatives to generate three oxidized methylcytosines, i.e. 5hmC, 5fC and 5caC. Highly oxidized cytosine bases, 5fC and 5caC, are enzymatically excised by Thymine DNA glycosylase (TDG), and resulting abasic sites are repaired by the base-excision repair (BER) pathway to regenerate unmodified C, completing the DNA demethylation process (5mC to C). In standard bisulfite sequencing (BS-seq), 5mC and 5hmC are resistant to sodium bisulfite-mediated deamination and read as C in subsequent sequencing, whereas unmodified C, 5fC and 5caC are read as T. M.SssI exhibits robust methylase activity toward unmodified cytosines within CpGs. In MAB-seq, only 5fC and 5caC are read as T after genomic DNA is treated with M.SssI. In caMAB-seq, genomic DNA is first treated with sodium borohydride (NaBH4) to reduce 5fC back to 5hmC, enabling 5caC to be directly mapped as T.