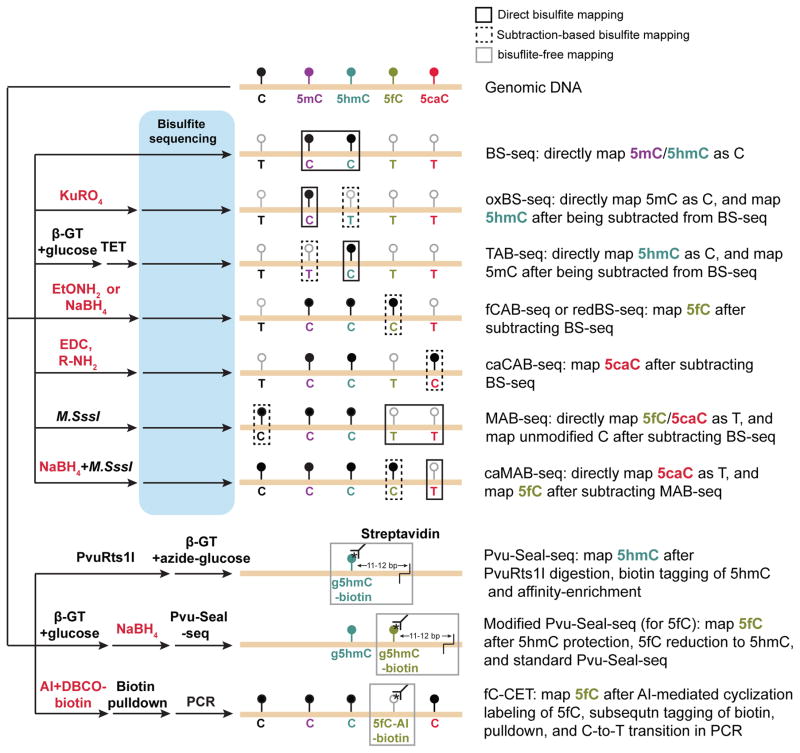
Figure 3. Schematic diagram of base-resolution mapping methods for oxidized methylcytosines.
Coupled with various chemical (in red) and enzymatic (in black) treatments, bisulfite sequencing (BS-seq) based (upper panels) or bisulfite-free (lower panels) base-resolution mapping methods have been developed to profile 5fC and 5caC. In direct bisulfite mapping (targeting modification highlighted in black line boxes), position and abundance of specific oxidized methylcytosines (e.g. mapping of 5hmC by MAB-seq 46) can be directly determined. In subtraction-based bisulfite mapping (targeting modification highlighted in dash line boxes), subtracting signals between conventional BS-seq and those of modified BS-seq (e.g. mapping of 5hmC by subtracting signals of oxBS-seq 67 from those of standard BS-seq) is required to indirectly determine the position and abundance of oxidized methylcytosines. In two bisulfite-free mapping strategies (targeting modification highlighted in gray line boxes), modification-sensitive restriction enzyme (5hmC-specific endonuclease PvuRts1l for Pvu-Seal-seq) or chemical (AI [azido derivative of 1,3-indandione] for fC-CET) assisted tagging of oxidized methylcytosines is utilized to both enrich the genomic fragments containing methylcytosines (giving relative abundance) and to identify the position of these cytosine variants.