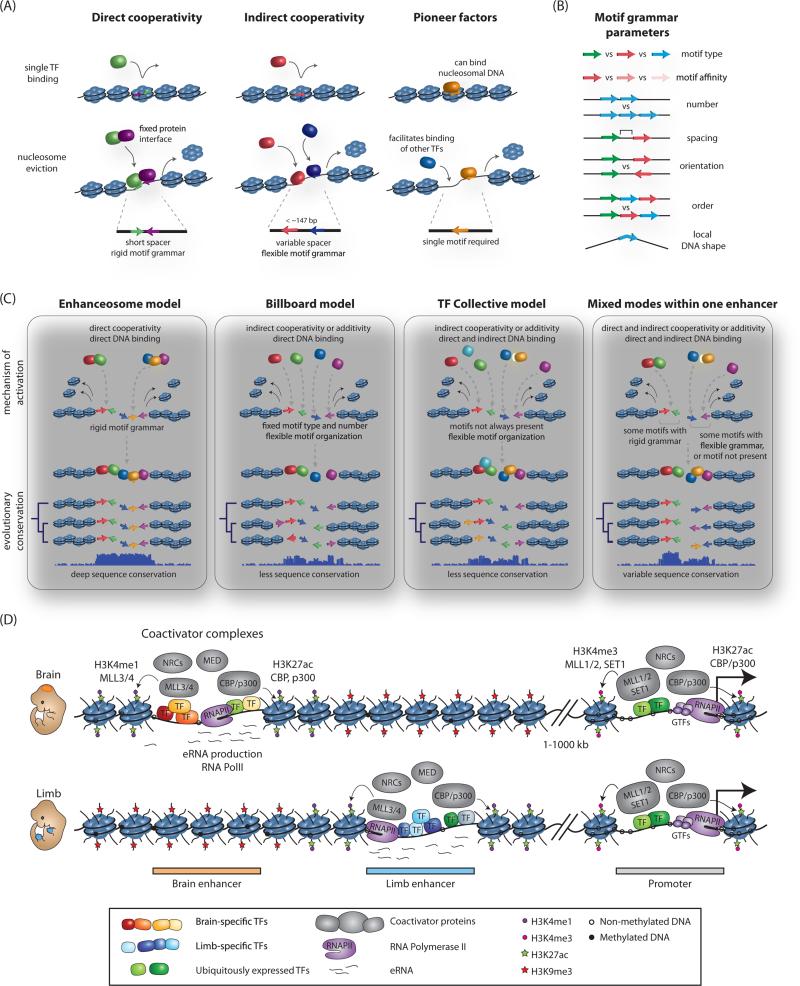
Figure 1. Nucleosome eviction, enhancer grammar and models of enhancer architecture.
(A) TF mechanisms for overcoming energetic barrier to nucleosomal eviction and underlying motif requirements. (B) Parameters of motif grammar at enhancers. (C) Models of enhancer architecture and their primary mechanism of TF binding, flexibility of motif organization, and selective constraint across evolution. (D) Coactivator binding and chromatin signatures at active enhancers. Presence of cell-type specific and broadly-expressed TFs, RNA Polymerase II (RNAPII) and associated enhancer RNAs (eRNAs), coactivator complexes such as Mediator (Med), nucleosome remodeling complexes (NRCs), histone acetyltransferases (CBP/p300) and methyltransferases (MLL3/4) at tissue-specific enhancer elements is schematically depicted for a brain (top) or limb (bottom) specific enhancer. Select modifications of neighboring nucleosomes associated with active enhancer states are highlighted. An overlapping set of protein complexes and modifications is also present at promoters, with the distinction that enhancers have high ratio of H3K4me1 to H3K4me3, and the reverse is true at promoters. Both active enhancers and promoters are characterized by DNA hypomethylation, with the methylation status of enhancers being more dynamic and tracking with their cell-type specificity.