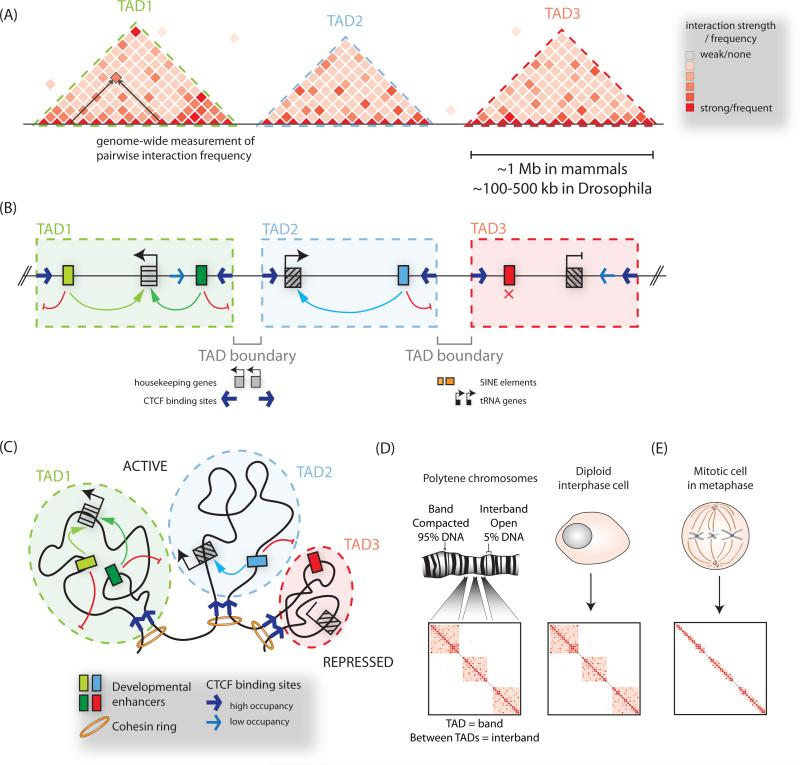
Figure 3. Organization of chromatin into topologically associated domains.
(A) Hi-C or 5C heatmaps visualize three-dimensional interactions, or compartmentalization of chromosomes into topologically associated domains (TADs), visible as triangular blocks of increased interaction frequencies. (B) TAD boundaries restrict the influence of regulatory elements to genes within a given TAD, and limit the spread of chromatin modifications. The boundary regions between TADs have been observed to be associated with CTCF binding sites, housekeeping genes, SINE elements and tRNA genes. (C) A model of chromosome folding corresponding to (B), with convergent CTCF sites and cohesin depicted at loop anchors. (D) Correspondence between TADs and cytological bands on polytene chromosomes, and TAD boundaries with decondensed interbands. (E) During mitosis TAD structure is lost.