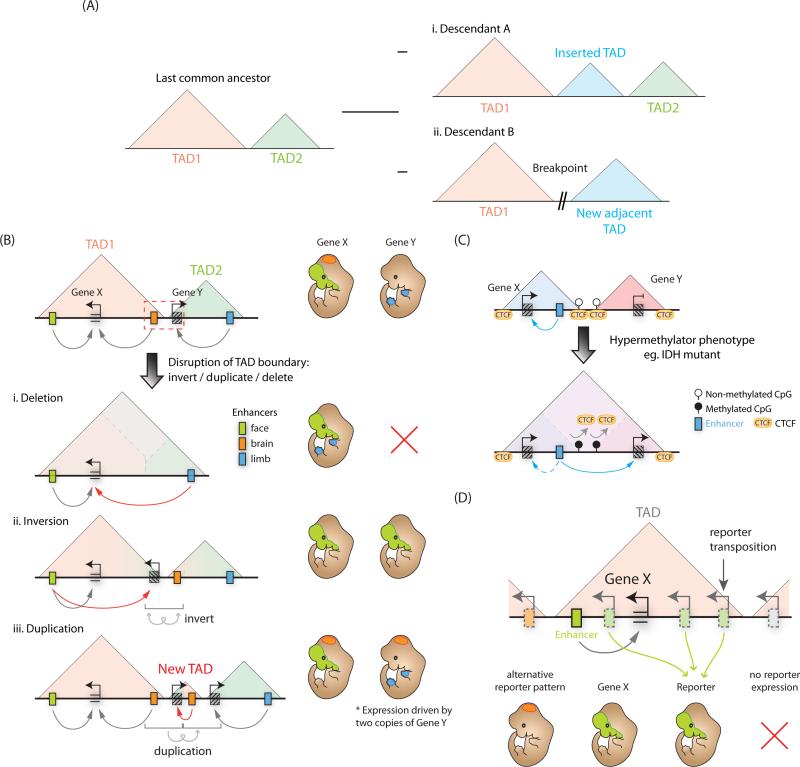
Figure 4. Topologically associated domains define discrete units of gene regulation.
(A) TAD boundaries are highly conserved across mammalian evolution, therefore structural changes tend to occur at TAD boundaries, for example (i) insertion of a new TAD or (ii) chromosomal breaks. (B) Disruption of a TAD boundary (red, dashed box) can impact gene expression. (i) Boundary deletion fuses two adjacent TADs facilitating de novo regulation of genes from one TAD by the enhancers in another. (ii) Boundary inversion can translocate genes or enhancers into an adjacent TAD where they are then incorporated into the regulatory environment of the new TAD. (iii) TAD boundary duplication can create a new TAD and potentially expose any duplicated genes to a novel regulatory environment. (C) CTCF binding sites can be DNA methylation sensitive, therefore aberrant DNA methylation can abrogate CTCF binding, causing TAD boundary defects and misregulation of gene expression. (D) Reporter constructs transposed into different regions of a TAD containing a developmental enhancer, but not into genomic regions outside the TAD, can recapitulate gene expression pattern of the endogenous gene (Gene X) controlled by this enhancer.