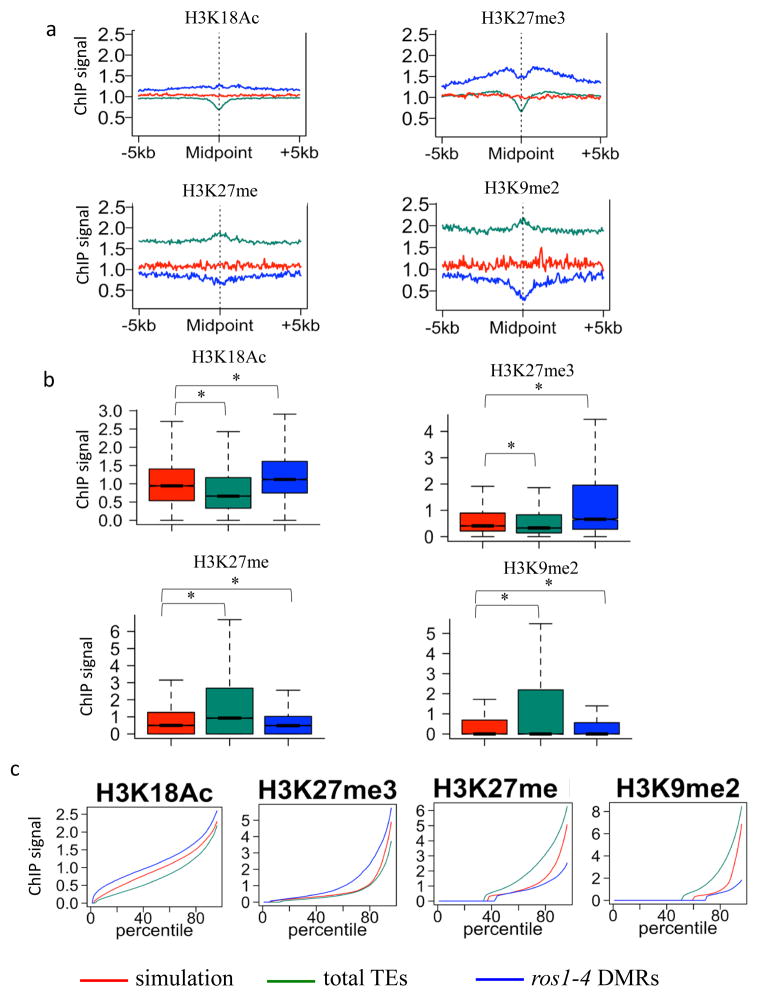
Figure 2. Chromatin features associated with ROS1 targets.
a. Association of different histone modifications surrounding ros1-4 hyper DMRs. Association of histone modifications at total TEs and simulated regions served as controls. In contrast to total TEs, ros1-4 hyper DMRs are positively associated with H3K18Ac, H3K27me3, and negatively associated with H3K27me and H3K9me2.
b. Box plots display of the results in Fig. 2a (*p-value < 1e-15, one-tailed Wilcoxon rank sum test).
c. Percentile plots of the same data as in Fig. 2b. For each histone mark, simulated regions, TEs and ros1-4 DMRs were ranked based on their histone ChIP signals from low (left) to high (right) along X-axis. X-axis is ranking percentile, and Y-axis is ChIP signal.