Figure 2.
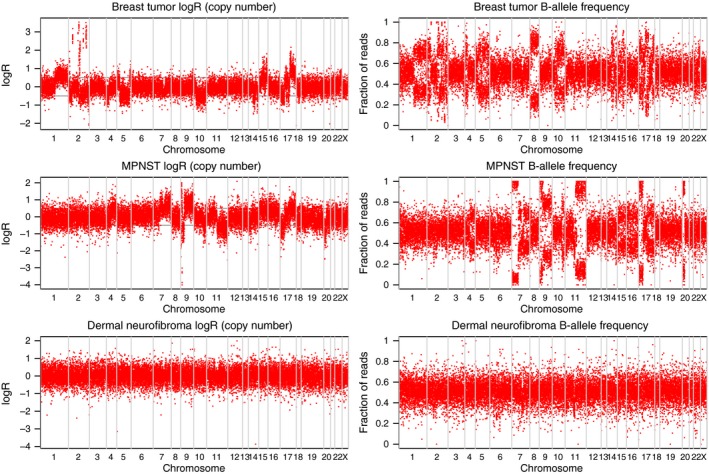
Analysis of copy number alterations in breast cancer (A), malignant peripheral nerve sheath tumor (MPNST) (B) and dermal neurofibroma (C), based on paired tumor and nontumor DNA analysis of exome sequencing data. Each red dot represents a genomic coordinate that was heterozygous in the germline sample. The breast cancer shows the most chromosomal rearrangements while the dermal neurofibroma shows none. Regions of allelic imbalance that also show a decrease in Log R typically represent Loss‐of‐heterozygosity (LOH); regions of allelic imbalance with no change in Log R show Copy‐Number Neutral LOH, and regions of allelic imbalance that correspond with increased Log R correspond to either focal amplifications (for example, several loci in chromosome 2 of the breast cancer) or large‐scale amplifications. The changes in Log R and allele frequency observed across multiple chromosomes in breast cancer and MPNST indicate allelic imbalance or copy number alterations in these samples, but not in the dermal neurofibroma.
