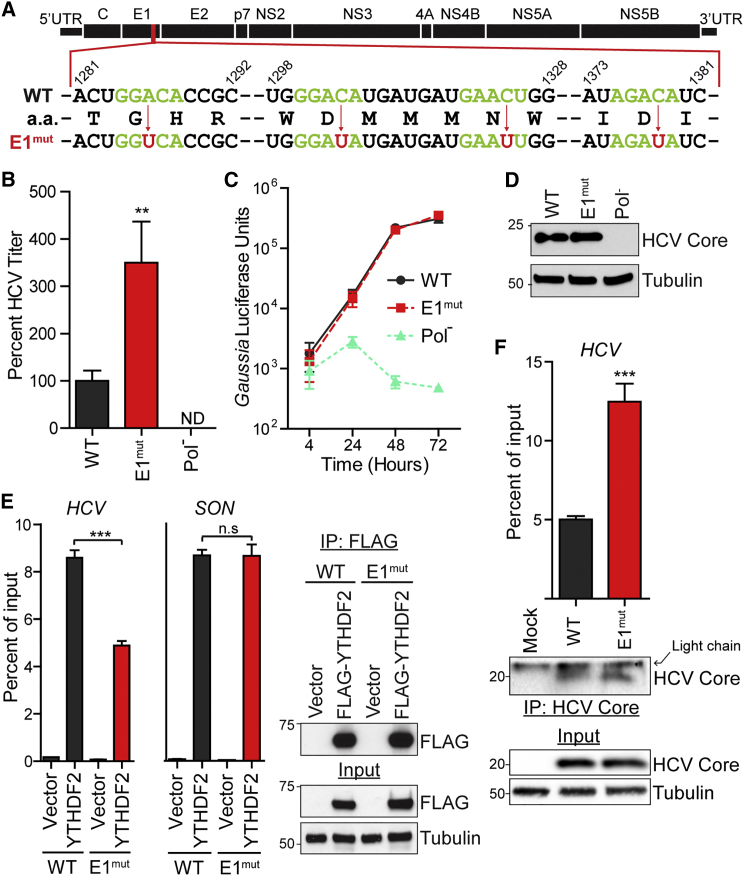
Figure 5.
m6A-Abrogating Mutations in E1 Increase Infectious HCV Particle Production
(A) Schematic of the HCV genome with the mutation scheme for altering A or C residues (red arrows) to make the E1mut virus. Consensus m6A motifs (green) and inactivating mutations (red) are shown. Dashes represent nucleotides not shown. Genomic indices match the HCV JFH-1 genome (AB047639).
(B) FFA of supernatants harvested from Huh7 cells after electroporation of WT or E1mut HCV RNA (48 hr) and analyzed as the percentage of viral titer relative to WT.
(C) Gaussia luciferase assay to measure levels of the WT, E1mut, or Pol− HCV luciferase reporter (JFH1-QL/GLuc2A) transfected in Huh7.5 CD81 KO cells.
(D) Immunoblot analysis of extracts of WT, E1mut, or Pol− JFH1-QL/GLuc2A transfected in Huh7.5 CD81 KO cells.
(E) Enrichment of WT or E1mut reporter RNA or SON mRNA by immunoprecipitation of FLAG-YTHDF2 or vector from extracts of Huh7 cells. Captured RNA was quantified by qRT-PCR and graphed as the percentage of input. Right: immunoblot analysis of anti-FLAG immunoprecipitated extracts and input.
(F) Enrichment of WT or E1mut HCV RNA by immunoprecipitation of HCV Core from extracts of Huh7 cells electroporated with the indicated viral genomes (48 hr). Lower: immunoblot analysis of anti-Core immunoprecipitated extracts and input.
Data are representative of two (D and E) or three (B, C, and F) experiments and presented as the mean ± SD (n = 3). ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001 by unpaired Student’s t test. See also Figure S5.