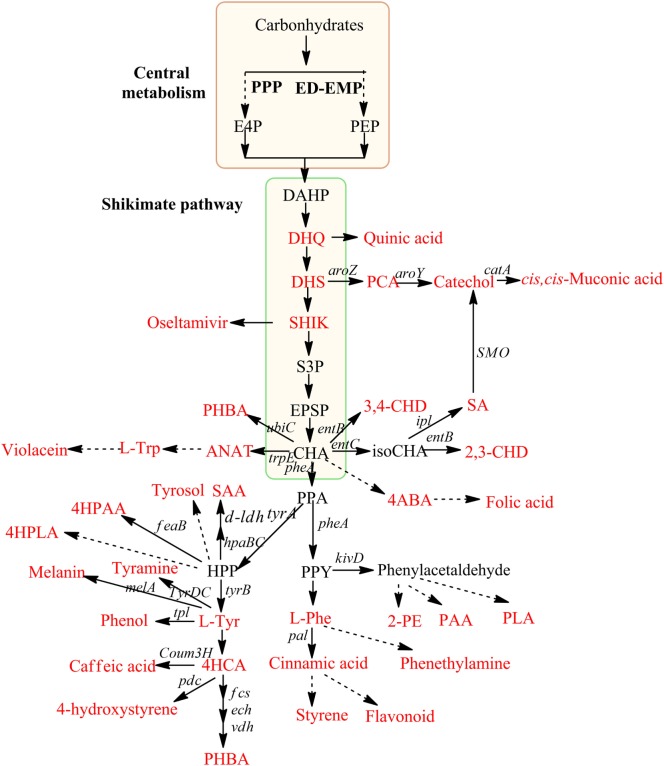
Figure 1.
Production of aromatic compounds via shikimate pathway in microbes. Red framed box indicates central metabolism, and green framed box indicates shikimate pathway. Compounds have industrial interest are highlighted in red color. Dashed arrows indicate multi-enzymatic synthesis. DHQ, 3-dehydroquinate; DHS, 3-dehydroshikimate; SHIK, shikimate; S3P, shikimate-3-phosphate; EPSP, 5-enolpyruvyl-shikimate 3-phosphate; CHA, chorismate; HPP, 4-hydroxy phenyl pyruvate; ANAT, anthranilate; PPA, prephenate; PPY, phenyl pyruvate; 4ABA, 4-aminobenzoic acid; SAA, salvianic acid A; 4HCA, p-hydroxy cinnamic acids; PLA, phenyl lactic acid; 4HPLA, 4-hydroxy phenyl lactic acid; 2-PE, phenyl ethanol; PAA, phenyl acetic acid; 4HPAA, 4-hydroxy phenyl acetic acid; 2,3-CHD, S,S-2,3-dihydroxy-2,3-dihydrobenzoic acid; 3,4-CHD S,S-3,4-dihydroxy-3,4-dihydrobenzoic acid; aroZ, 3-DHS dehydratase; aroY, PCA decarboxylase; catA, catechol 1,2-dioxygenase; feaB, phenylacetaldehyde dehydrogenase; tpl, Tyr phenol lyase from Pantoea agglomerans; tyrDC, tyramine decarboxylase gene from Lactobacillus brevis; pdc, 4HCA decarboxylase from L. plantarum; tyrB, tyrosine aminotransferase; hpaBC, an endogenous hydroxylase from E. coli; ldh, lactate dehydrogenase from L. pentosus; pal, Phe-ammonia lyase/Tyr-ammonia lyase from Rhodosporidium toruloides; kivD, phenylpyruvate decarboxylase tyrA, biofunctional chorismate mutase/prephenate dehydrogenase; entB, isochorismatase; entC, isochorismate synthase; melA, tyrosinase; fcs, p-coumaroyl-CoA synthetase; ech, p-coumaroyl-CoA hydratase/lyase; vdh, p-hydroxybenzaldehyde dehydrogenase; ipl, isochorismate pyruvate lyase; SMO, salicylate 1-monoxygenase; pheA, bifunctional chorismate mutase/prephenate dehydratase; tal, tyrosine ammonia lyase; Coum3H, 4-coumarate hydroxylase; ubiC, chorismate.