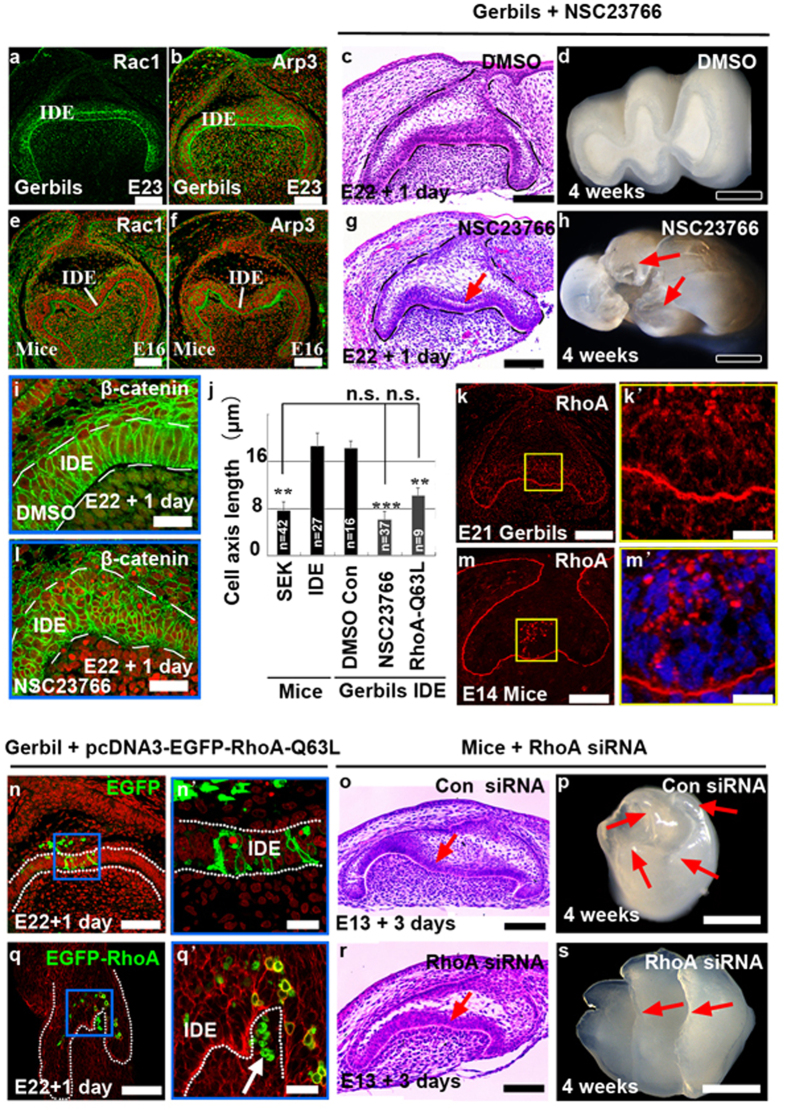
Figure 3. Rac1 and RhoA mediate epithelial invaginations and cuspal shapes.
(a,b,e,f) Localization of Rac1 and Arp3 in the first molar in gerbils and mice at the bell stage. (c,d,g,h) Phenotypes of gerbil tooth germs and teeth after treatment with the DMSO and NSC23766 (90 μM in DMSO). The arrow in panel g indicates ectopic invagination of IDE, and the arrows in panel h indicate cusps. (i,l) Localization of β-catenin in the cuspal regions in the tooth germ with or without NSC23766 shows cell shapes and sizes. TO-PRO-3 was used as counter staining. (j) The cell length of the SEK in mice and IDE cells in gerbils with NSC23766 or pcDNA-EGFP-RhoA-Q63L were dramatically shortened when compared with the length of the IDE in mice and in gerbil controls (**P ≤ 0.01; ***P ≤ 0.001). n.s., not significant. The error bars indicate SM. (k,m) Localization of RhoA in the first molar in gerbils and mice at the early cap stage. (n,n’,q,q’) The formation of a pit (q’; arrow) and changes of cell shapes after electroporation of pcDNA-EGFP-RhoA-Q63L (q,q’) compared to electroporation with pcDNA-EGFP vehicles (N, N’) in IDE cells. (n’,q’) Higher magnification images of the blue boxes in panel n and q. (o,p,r,s) Phenotypes of tooth germs (o,r) and teeth (p,s) after treatment with control siRNA and RhoA siRNA (300 nM). Arrows in panel o and r show the loss of invagination of IDE by RhoA siRNA. Arrows in panel p and s indicate the loss of cusps and the formation of ridges by RhoA siRNA. Stages are indicated in each panel. IDE, inner dental epithelium. Scale bar, 500 μm (d,h,p,s), 200 μm (n,q), 100 μm (a–c,e–g,k,m,o,r), 20 μm (i,l,k’,m’,n’,q).