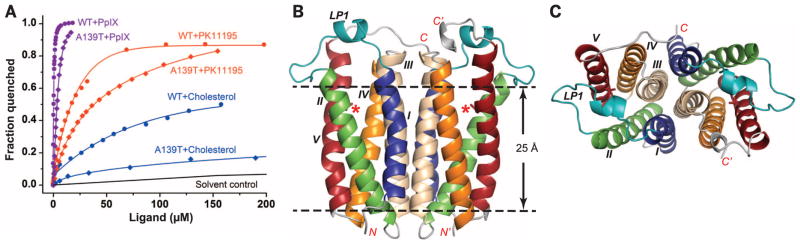
Fig. 1. Structure and ligand binding affinity of RsTSPO WT and the A139T mutant.
(A) Ligand binding affinities are shown for RsTSPO WT and the A139Tmutant mimicking the human A147Tmutation. Dissociation constant (Kd) values are obtained as described in (24): 10 ± 1 μM for PK11195 with WT, 42 ± 4 μM with A139T; 0.3 ± 0.01 μM for PpIX with WT, 1.9 ± 0.3 with A139T; and ~80 μM for cholesterol with WT, >300 μM with A139T. WT data are from (14), reproduced for comparison. (B) Overall structure of the A139T dimer. The position of the A139T mutation is labeled with a red asterisk and shown in sticks; the five transmembrane helices (TM-I to TM-V) are colored blue, green, wheat, orange, and red, respectively; and loop 1 (LP1) is colored teal. (C) Top view of (B). RsTSPO A139T crystallized in two different space groups (C2 and P212121) that have identical overall structures except for the flexible C terminus, whereas WTcrystallized in a P21 space group. In all three crystal forms, the identical parallel dimer of RsTSPO was observed (Fig. 2). The A139Tmutant in the C2 space group is shown here and used to discuss the major structural features of RsTSPO, as it has the highest resolution and most complete structure of RsTSPO. The N and C termini are labeled N/N′ and C/C′, respectively; the dashed lines highlight the approximate membrane region.