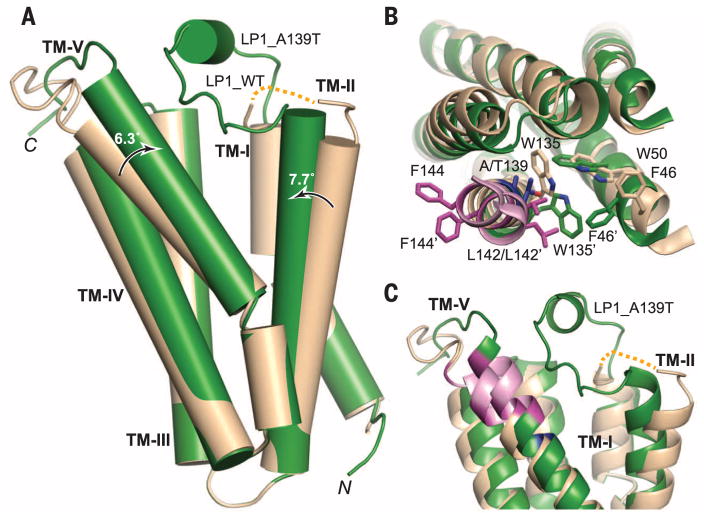
Fig. 3. Structural comparison of WTand A139T RsTSPO.
Monomers of the WT (wheat) and A139T (green) are overlaid. (A) Overall structural alignment that highlights the degree and direction (arrows) of conformation change from WT to A139T for TM-II and TM-V. (B) Top view of the monomer, highlighting the side-chain rearrangements in sticks. Residue 139 is colored in blue; the CRAC site is colored in pink with two of three proposed critical residues (L142 and F144) highlighted in magenta. The prime symbol designates the mutant position. (C) Close-up side view of the potential ligand binding cavity, which reveals major differences in the conformations of LP1, TM-II, and TM-V between the WTand mutant proteins (fig. S7). The dotted yellow line denotes the location of unresolved LP1 in the WT structure (residues 29 to 40).