Figure 1. Methods for probing RNA secondary structure.
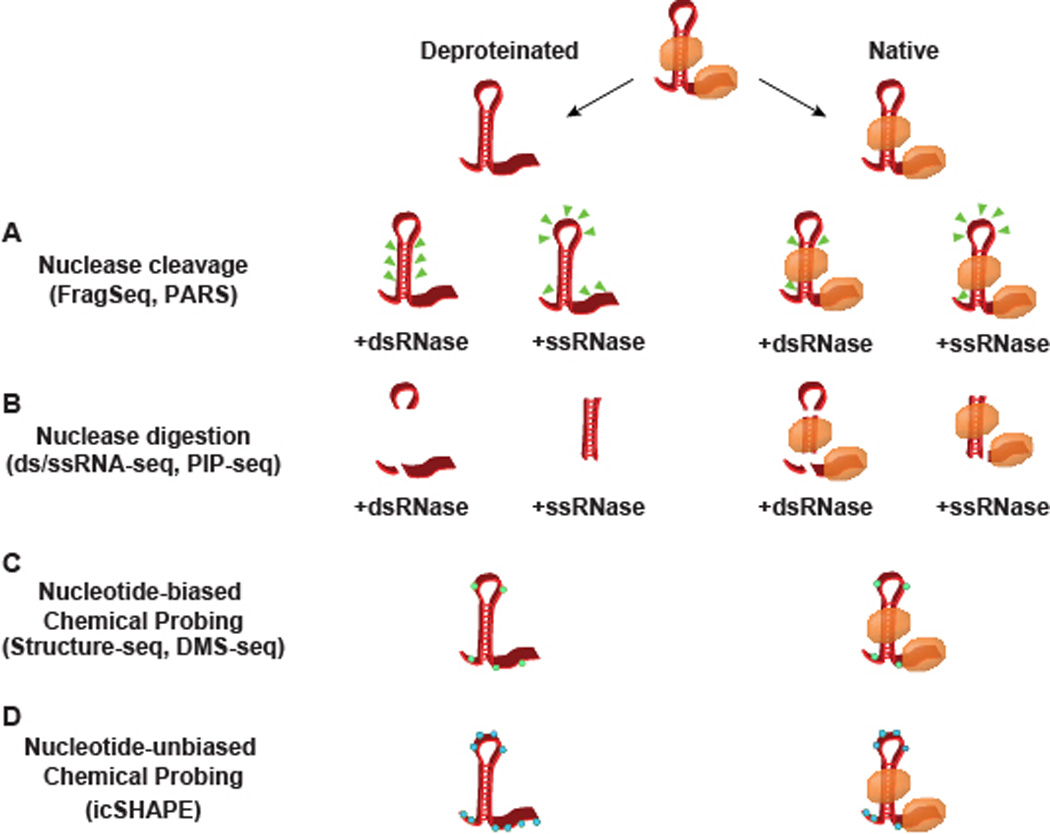
A schematic representation of the nuclease- and chemical-based probing techniques for empirically determining secondary structure. RNA can either be probed in a native state bound by RNA binding proteins (orange ovals) or deproteinated through extraction protocols or proteinase K treatement. (A) PARS assigns structure by the sites of transcript cleavage (green triangles), whereas (B) dsRNase/ssRNase-seq and PIP-seq both work by complete digestion. (C–D) Chemical probing works through reagents that preferentially adduct to nucleotides in a single-stranded confirmation, forming covalent modifications in either a (C) nucleotide-biased (green hexagons) or (D) unbiased (blue hexagons) manner. While multiple cleavage sites and covalent modifications are represented in this schematic, it is worth noting that PARS and the chemical probing techniques work with single-hit stoichiometry, with one cut/modification site interrogated per sequencing read.
