Abstract
Extrapulmonary tuberculosis (TB) is a significant public health challenge in South Africa and worldwide, largely fuelled by the HIV epidemic. In spinal TB, Mycobacteria infect the spinal column without dissemination to the spinal cord. The immune microenvironment, target cell characteristics, and other evolutionary forces within granulomas during HIV/TB coinfection are poorly characterized. We investigated whether spinal TB granulomas represent a sequestered anatomical site where independent HIV evolution occurs, and assessed the role of macrophages as a target cell for both HIV and mycobacteria. RNA was extracted from plasma and granulomatous tissue from six antiretroviral-naive HIV-1/spinal TB-coinfected patients, RT-PCR amplified, and the C2-V5 env segment was cloned and sequenced. Analysis of genetic diversity, phylogeny and coalescence patterns was performed on clonal sequences. To investigate their role in HIV sequestration, macrophages and the HIV-1 p24 protein were immune localized and ultrastructural features were studied. Intercompartment diversity measurements and phylogenetic reconstruction revealed anatomically distinct monophyletic HIV-1 clusters in four of six patients. Genotypic CCR5-tropic variants were predominant (98.9%) with conservation of putative N-linked glycosylation sites in both compartments. CD68+ reactivity was associated with higher tissue viral load (r = 1.0; p < 0.01) but not greater intrapatient diversity (r = 0.60; p > 0.05). Ultrastructural imaging revealed the presence of bacterial and virus-like particles within membrane-bound intracellular compartments of macrophages. Spinal tuberculosis granulomas may form anatomically discreet sites of divergent viral evolution. Macrophages in these granulomas harbored both pathogens, suggesting that they may facilitate the process of viral sequestration within this compartment.
Introduction
One of the most devastating complications of the worldwide HIV-1/AIDS epidemic has been the dramatic increase in the incidence of active tuberculosis (TB) at pulmonary and extrapulmonary sites.1 HIV infection contributed to 1.1 million new TB infections and approximately 320,000 TB-related deaths in 2012 alone; 75% of these occurred in Africa.1 This enormous disease burden represents a serious challenge to TB control programs in resource-constrained regions where access to combination antiretroviral therapy (ART) and large-scale antituberculosis prophylaxis is still suboptimal and logistically challenging. HIV-1-induced immunodeficiency leads to increased risk of newly acquired TB and adverse clinical outcomes.2 Furthermore, HIV replication, diversity, persistence in tissue compartments and reservoirs complicate clinical outcomes and pose a threat to therapeutic management and cure strategies.3,4
HIV-1 diversity is a function of its error-prone reverse transcriptase, replication rate, recombination, and host selective pressures.5 Following transmission, HIV-1 spreads to various anatomical sites where the site-specific microenvironment regulates its evolutionary kinetics.6,7 HIV-1 replication in remote or sequestered anatomical sites may have clinical consequences by creating a source of virus for transmission, drug resistance (due to poor drug penetration), tissue-specific clinical deterioration, and establishment of long-lived viral reservoirs.8,9 The role of coinfections such as TB, and related immune and cellular microenvironment remodeling in promoting independent viral evolution at tissue sites of infection, is poorly understood. We do know that these sites have clinical relevance as they may prevent access of infected cells to ART and appropriate host immune responses.3 Anatomical compartments representing discrete sites of independent viral evolution include the central nervous system, genital tract, kidney epithelium, and lymph nodes.10–14 Whether the same is true within TB coinfected granulomas is currently unclear and inconclusive.15–17 Understanding the dynamics of viral evolution within this unique highly compartmentalized but immunologically active environment is important to virus eradication strategies. The nature of the viral variants, their response to treatment, and the mechanisms that govern their progression to sequestration may play critical roles in clinical patient management.
The host immune response to TB is not fully characterized but involves primarily macrophages and CD4+ and CD8+ T cells.2,18 Histopathology studies of pulmonary and pleural TB have been performed18 but exploratory investigations of the spine are rare.19,20 None of these studies, however, fully explores the role of specific cell types, such as macrophages, as drivers and/or sites of divergent HIV evolution.
In this study we sought to investigate whether the extradural spinal TB granuloma formed an anatomical site of HIV sequestration by analyzing clonal sequences generated from the blood and spinal granuloma biopsies of coinfected patients. Our investigation revealed evidence of divergent viral evolution within spinal TB granulomas. We also explored the immune microenvironment with specific focus on macrophages given that TB and HIV are intracellular pathogens that target macrophages. Immunolocalization studies revealed the presence of HIV-infected macrophages while electron microscopy provided evidence of HIV and TB particles within an intracellular endosomal compartment.
Materials and Methods
Study subjects, clinical assessment, and specimen collection
The Institutional Review Board of the University of KwaZulu-Natal approved the study protocol (Ref. #H112/02). All participants or their legal guardians provided informed consent. Blood and matched granuloma tissue were obtained from six HIV-1/spinal tuberculosis coinfected patients undergoing vertebral excision and anterior spinal decompression to resolve associated neurological deficits at King George V Hospital (now King Dinuzulu Hospital) in Durban, South Africa. Given that extrapulmonary TB with HIV coinfection is AIDS defining (WHO stage 4) all patients were classified accordingly. Cerebrospinal fluid (CSF) was not sampled since there was no clinical justification to sample the CSF given that the Mycobacterium tuberculosis (Mtb) infection had not disseminated and breached the blood–brain barrier. All patients received preoperative nutritional support to achieve safe hemoglobin and albumin levels (>100 g/liter and >300 g/liter, respectively) and standard antituberculosis therapy (600 mg rifampicin, 400 mg isoniazid, 1,500 mg pyrazinamide, 1,500 mg and 1,200 mg ethambutol daily) for a mean period of 8 weeks (range = 5–24 weeks). All patients were antiretroviral (ARV) drug naive since ART had not been rolled out as part of the national policy at the time of sample collection.
The excised granulomatous tissue was processed immediately for histology, bacterial culture diagnostics, and molecular assays. A venous blood sample was collected for quantification of HIV-1 viral load (Nuclisens HIV-1 QT kit, Organon Teknika) and T-lymphocyte subsets (Beckman Coulter TetraOne method). For tissue viral load analysis (Nuclisens HIV-1 QT kit), HIV RNA was extracted from 10 mg of tissue. Tissue was weighed and then homogenized in lysis buffer (Nuclisens isolation kit, Biomerieux) prior to RNA extraction using the MiniMag Magnetic Extraction protocol (Biomerieux) and HIV RNA was quantified with the Nuclisens HIV-1 QT Kit (Biomerieux). Viral loads were expressed as cp/ml of blood for plasma samples and cp/mg of tissue in the case of the granulomas.
Immunolocalization of CD68+/p24+ cells
CD68+ cells (macrophages/monocyte lineages; Clone KP1, Dako; dilution 1:200) and HIVp24 antibody (Clone Kal-1, Dako, Denmark; dilution 1:10) were immunolocalized to 5-μm-thick sections of granulomatous tissue using an indirect immunoperoxide-staining protocol (Dako Envision Dual Staining kit, Dako). Visualization and capture of positive staining cells utilized the Soft Imaging System (SIS). The region of interest in each section at the same initial magnification (×40) was selected based on typical features of a granuloma, namely, a central core of necrosis containing macrophages and multinucleated giant cells surrounded by lymphocytes. Additional biopsies were fixed, processed, and embedded using conventional techniques for transmission electron microscopy (TEM). Ultrathin sections (50 nm) were stained according to standard protocols and viewed with the Jeol JEM-1011 transmission electron microscope and iTEM imaging software.
PCR amplification and cloning
Paired plasma and tissue samples were available for sequencing for six patients (three adults, three children). HIV-1 RNA templates, extracted using the MiniMag extraction protocol, were reverse transcribed (Superscript III reverse transcriptase; Invitrogen, San Diego, CA) and a 621-bp segment of the env gene (7,026 to 7,647 by HXB2 numbering) was amplified as previously described.21 Purified amplicons (Qiagen, Valencia, CA) were cloned into the pCR2.1-TOPO TA vector (Invitrogen) and positive plasmids (5–10 clones) were purified and sequenced (Big-Dye terminator V3.1).
Sequence editing, alignment, and analysis
All HIV-1 env choromatograms were imported into Geneious v6 (www.geneious.com), visually edited, and accepted for analysis if quality scores were >90%. To exclude potential contamination, newly derived sequences were compared to subtype reference strains (Los Alamos database; http://hiv-web.lanl.gov/content/hiv-db/SUBTYPE_REF/align.html) and to HIV-1 sequences previously amplified in our laboratory. Consensus sequences generated in Geneious were codon aligned in Clustal W22 and manually edited in SeAL (http//:tree.bio.ed.ac.uk). Phylogenetic trees were constructed for the entire dataset against HIV-1 reference sequences and for the plasma and granuloma sequences from individual patients. Maximum likelihood (ML) and Bayesian trees were reconstructed in RaxML23 and MrBayes24 respectively using appropriate models estimated in ModelTest25 and an estimated gamma heterogeneity alpha parameter. The ML tree topology was optimized in RaxML and the reliability of the internal nodes was estimated using 1,000 bootstrap replicates (ML trees) or four Markov Chain Monte Carlo (MCMC) chains with 106 generations and a 10% burning (Bayesian trees). Results were visualized using Tracer parameters where an effective sample size (ess) >200 suggested good mixing and sampling of the trees. Consensus trees and posterior support for internal nodes were generated in TreeAnnotator (www.beast2.org/wiki/index.php/TreeAnnotator) and annotated in FigTree (http://tree.bio.ed.ac.uk/software/figtree/).
Analysis to confirm viral compartmentalization and migration
Assessment of viral compartmentalization was performed using tree-based and diversity-based algorithms in DIVEIN (http://indra.mullins.microbiol.washington.edu/DIVEIN/) and HyPhy.26 We performed the intracompartment diversity and the two-sample diversity tests (z-test and t-test) in DIVEIN. In Hyphy we performed two tree-based tests of compartmentalization: the Slatkin–Maddison (SM) test to calculate the probability of as many or fewer migrations from one compartment to the other occurring by chance, the branch count correlation coefficient (rb), and the branch length correlation (r). We also employed the distance-based Hudson nearest-neighbor test (S-nn; TN93 model, 500 resampling). All sequences were screened for signature patterns indicative of viral tropism (CCR5 or CXCR4 usage) using the online Geno2Pheno tool (www.geno2pheno.org/).27
Statistical analysis
All statistical analyses were undertaken in Stata v11. A 5% (p < 0.05) level of significance was accepted for all analyses unless otherwise stated.
Results
Clinical and demographic findings
All patients (n = 6) were diagnosed with active spinal TB based on presumptive clinical and radiological findings. Their demographic and clinical information are summarized in Table 1. Data on when these patients acquired their HIV infection were not available. Because patients did not present with symptoms suggesting other viral infections related diagnostic tests were not performed. Since mycotic infections of the spine have a clinical presentation similar to spinal TB, patient samples were tested for evidence of fungal infection and were found to be negative. Tissue viral loads were high and well in excess of 5 log/mg in all patients. There was no correlation between tissue and plasma viral load measurements (Table 1).
Table 1.
Demographic, Clinical Characteristics, and Basic Laboratory Findings of Study Population
| Patient Id. | Sex | Age (years) | CD4 count (cells/μl or %)a | CD8 count (cells/μl or %)a | Plasma viral load (log10 cp/ml) | Tissue viral load (log10 cp/mg) | Culture identification | Clinical detailsb |
|---|---|---|---|---|---|---|---|---|
| TB12 | Female | 2 | 21* | 40* | 4.2 | 8.1 | No growth | No neurology (Frankel E) |
| TB25 | Female | 3 | 23* | 50* | 4.3 | ND | No growth | Spastic paraplegia |
| TB32 | Male | 2 | 12* | 30* | 6.4 | 9.2 | Mtb | Advanced AIDS; oral thrush, enlarged and inflamed lymph nodes |
| TB16 | Male | 28 | ND | ND | 4.5 | 6.5 | No growth | Paraplegic; complete immobility (Frankel A); incontinent |
| TB20 | Female | 25 | 100 | 661 | 5.7 | 5.4 | Mtb | ND |
| TB38 | Female | 43 | ND | ND | 4.9 | 8.2 | Mtb | ND |
| Mean ± SD (range) | ||||||||
| Adults | 32 ± 15.9 | — | — | 5.0 ± 0.5 | 6.7 ± 1.1 | |||
| Children | 2.3 ± 2.1 | 18.6 ± 6.1* | 40 ± 16.2* | 4.9 ± 1.2 | 8.7 ± 0.8 |
CD4 and CD8 measurements for adults (≥16 years old) are presented as absolute values and for children (<16 years old) are presented as percentages as indicated by an asterisk(*).
Clinical data are presented where available.
ND, no data available; Mtb, Mycobacterium tuberculosis.
Phylogenetic tree-based analysis of divergence
A total of 94 sequences, generated from tissue and plasma (50 plasma-derived and 44 tissue-derived sequences) of six patients, were analyzed. Our objectives were first, to reconstruct patient sequences in relation to reference sequences, and second, to determine the clustering and gene flow patterns of sequences derived from the two anatomic compartments of individual patients. Phylogenetic reconstruction of clonal and published HIV-1 reference sequences (Subtypes A to K) confirmed that all patient sequences were HIV-1 subtype C (Fig. 1A). Tissue-derived and plasma-derived sequences from each patient clustered together, distinct from those of other participants, indicating that there was no interpatient contamination.
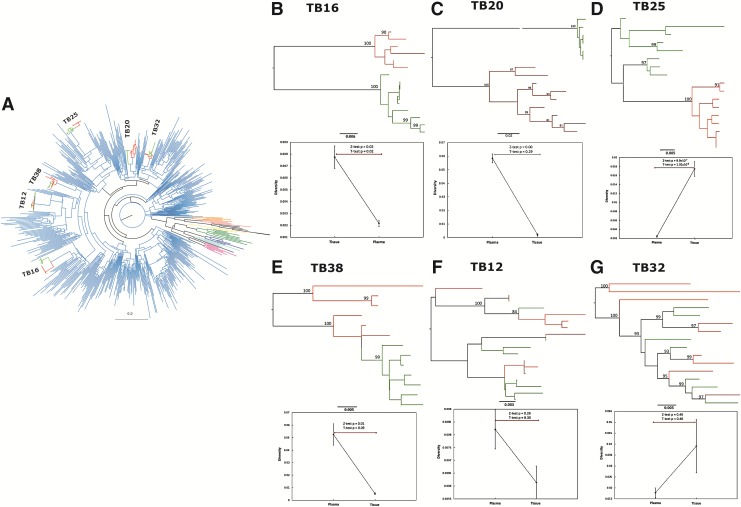
FIG. 1.
(A) A maximum likelihood tree of clonal plasma (red) and tissue-derived (green) sequences from individual patients clustering within the subtype C lineages (blue). Reference sequences of subtypes A (pink), B (purple), D (light blue), F (dark green), G (black), H (orange), J (salmon), and K (yellow) are also represented. The phylogenetic reconstruction was performed in RaxML using the HKY85 model with gamma distribution. (B–G) Phylogenetic reconstructions illustrating maximum likelihood trees constructed in RaxML of individual patient plasma-derived (red) and tissue-derived (green) sequences with branch support indicated are represented. (B–E) Two distinct patterns of clustering are observed where monophyletic clustering is shown [(B) TB16; (C) TB20; (D) TB25; (E) TB38)] and intermixing of variants is illustrated [(F) TB12 and (G) TB32]. The panels below each phylogenetic tree are graphic representations of intracompartment diversity measures as estimated in DIVEIN, which illustrates the median diversity and IQR. Two-sample diversity tests (T) were conducted and illustrated are p-values derived from z-tests and t-tests comparing intracompartment diversity of plasma-derived with tissue-derived sequences.
Phylogenetic reconstruction of individual patient sequences demonstrated two distinct clustering patterns, the first of discrete monophyletic clusters of tissue-derived and plasma-derived sequences, and the second of intermingling (Fig. 1B–G). In four of six patients (Fig. 1B–E), plasma-derived and tissue-derived sequences clustered in discrete branches. The longer branch lengths observed, coupled with high internal branch support (posterior probability scores >0.9 and bootstrap values >80%), strongly suggested divergent evolution within these compartments. Tree topologies were suggestive of a founder effect where a single viral variant entered and, over time, expanded into a unique monophyletic cluster within spinal granulomas. In these four cases, all computational tests of compartmentalization (SM, R, r-b, Snn, and intracompartment distance) concurred with statistical significance (p < 0.01; Table 2 and Fig. 1B–E). Together, these findings confirmed the separation of plasma-derived and tissue-derived sequences into distinct monophyletic populations in these patients (Fig. 1B–E).
Table 2.
Summary of Tests of Compartmentalization Comprising Tree-Based (*) and Diversity-Based (#) Algorithms
| Intracompartment diversity#,a | CCR5 usagef,g | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Patient number | Plasma (± SD) | Tissue (± SD) | p-value (z-test; t-test) | SM*,b | R*,c | r-b*,d | Hudson#,eS-nn | Plasma (no. and % clones) | Tissue (no. and % clones) |
| TB12 | 0.008 ± 0.001 | 0.006 ± 0.001 | 0.29; 0.30 | 4 × 10−5 | 0.002 | 0.002 | 0.262 | 9 (100) | 9 (100) |
| TB25 | 0.002 ± 0.001 | 0.017 ± 0.002 | 9.9 × 10−7; 1.02 × 10−6 | 6.7 × 10−5 | 0.002 | 0.002 | 0.000 | 8 (100) | 8 (100) |
| TB32 | 0.002 ± 0.0002 | 0.040 ± 0.012 | 0.46; 0.46 | 0.022 | 0.002 | 0.03 | 0.000 | 9 (100) | 4 (100) |
| TB16 | 0.002 ± 0.0002 | 0.008 ± 0.001 | 0.02; 0.02 | 0.0005 | 0.002 | 0.002 | 0.000 | 6 (100) | 9 (100) |
| TB20 | 0.058 ± 0.003 | 0.002 ± 0.001 | 0.00; 0.29 | 0.0003 | 0.002 | 0.002 | 0.000 | 6 (77) | 9 (100) |
| TB38 | 0.052 ± 0.008 | 0.005 ± 0.001 | 0.006; 0.09 | 0.001 | 0.004 | 0.002 | 0.000 | 6 (100) | 7 (100) |
Intracompartment pairwise distance (DIVEIN).
Slatkin–Maddison test (HyPhy).
Branch length correlation coefficient (r, HyPhy).
Branch count correlation coefficient (rb; HyPhy).
Hudson nearest neighbor (Snn; HyPhy).
Coreceptor usage is indicated as percentage number of sequences.
Coreceptor usage potential was analyzed using the online tool Geno2Pheno (www.geno2pheno.org) that uses sequence data and support vector machine (SVM) classifiers to predict coreceptor usage.
Tests of compartmentalization were executed in DIVEIN and HyPhy software packages indicating statistical support for divergence and anatomical compartmentalization.
Statistical significance is indicated by p values where values <0.05 are underlined and <0.005 appear in bold.
The remaining two datasets, both from children <5 years of age, illustrated the second clustering pattern where plasma-derived and tissue-derived sequences were intermingled with unrestrained migration of HIV-1 variants between compartments (Fig. 1F and G). The tests of compartmentalization for these two study participants (Table 2; Fig. 2E and F) confirmed our visual observations in the phylogenetic trees; we therefore concluded that there was intermixing in these patients.
FIG. 2.
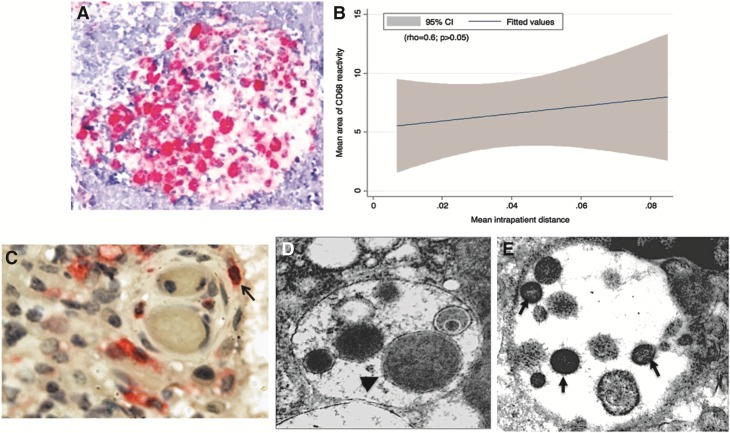
(A) A micrograph image of a typical granuloma illustrating the region of interest used for our analysis of CD68 immunolocalization (initial magnification, ×40). CD68-positive cells appear in pink and nuclei in blue. This image illustrates a typical granuloma with positively stained (pink) macrophages counted for semiquantitative analyses of intensity and area of immunolocalization. (B) A graphic illustration of the association (rho = 0.6; p > 0.05; 95% CI) between the area of CD68 immunoreactivity and the overall mean intrapatient genetic distance of plasma-derived and tissue-derived sequences. (C) A light micrograph and (D, E) transmission electron micrographs (TEM) of the spinal TB granulomas. Localization of macrophages (pink) and HIV p24 (black) appear in (C) (initial magnification ×40) with colocalized CD68+/p24+ within the same cells indicated by an arrow. (D) A TEM showing a bacterial cell (arrowhead) within an intracellular membrane-bound compartment. (E) Virus particles (arrows) were also observed, under TEM, within a similar membrane-bound intracellular compartment of a cell displaying macrophage morphology [(D, E) initial magnification ×50].
Analysis of viral tropism
Since key viral determinants of cell tropism and coreceptor usage are located within the env V3 loop,28 we investigated whether there were V3 loop amino acid differences between plasma-derived and tissue-derived sequences (Table 2). The biochemical properties (charge, size, hydrophobicity) at the amino acid positions determining the folding patterns of the V3 loop were similar and indicative of predominantly CCR5-utilizing variants in plasma and tissue. Only two plasma sequences from TB20 differed, where a 2-amino acid insertion resulted in a shift in putative coreceptor usage from CCR5 to CXCR4 (Table 2).
Immunohistology and ultrastructural morphology
Macrophages play an important role, both as target cells and mediators of the immune response, in HIV and TB disease pathogenesis. We therefore explored their possible role in the complex host–pathogen interaction within the anatomically remote spinal granuloma (Fig. 2A and B). While the overall number of CD68+ cells infiltrating the granulomas (indicated by the area of reactivity) was comparable, the level of CD68 expression (indicated by the intensity of the stain) differed and correlated with greater tissue viral load (rho = 1.0; p < 0.01) and greater intrapatient genetic distance (rho = 0.60; p = 0.2; Fig. 2B). These results suggest that increased site-specific macrophage activity is associated with higher HIV replication rates and viral loads in spinal TB-granulomas, although a causal relationship cannot be established.
For three of the six patients enough tissue samples were available for further IHC analysis of HIV and macrophage colocalization. Analysis of CD68+ cells and HIV-1 p24 antigen by immunohistochemistry within granulomas revealed that in three of the six patients selected for sequence analysis (TB12, TB 16, and TB 25) there was clear evidence of HIV p24-CD68 colocalization (Fig. 2C), with two of these patients (TB 16 and TB 25) possessing tissue-compartmentalized viral sequences. Finally, our investigations of ultrastructural features of coinfection and pathology revealed, under TEM, bacterial (Fig. 2D) and virus-like particles (Fig. 2E) within membrane-bound intracellular compartments in cells identified as macrophages.
Discussion
HIV-1-induced immune suppression has led to an increase in disseminated mycobacterial infections worldwide. Additionally, anatomical and cellular HIV reservoirs are of great clinical interest because they may impact pathogenesis, ART-mediated virus suppression, or strategies for HIV eradication.29 In this study we investigated the impact of anatomic sequestration of HIV, within a tuberculous spinal, granuloma on viral evolution. Using phylogenetic reconstruction and computational measures of compartmentalization30 we provide the first evidence to suggest divergent HIV evolution in spinal TB coinfected granulomas. These findings were supported by the fact that we noted no correlation between tissue and plasma viral load measurements, which suggests that viral variants in the tissue either accumulated slowly over time independent of plasma populations or are undergoing rapid tissue-specific expansion. We further demonstrated a possible role of macrophages as an intracellular compartment for viral sequestration and divergent evolution at the site of coinfection.
Our findings parallel those of Collins et al.15 that demonstrated divergent HIV evolution in pleural TB granulomas. In both instances, physical separation appears to promote viral evolution within TB-infected tissue. Unlike pulmonary tissue where migration of immune cells, both HIV infected and uninfected, is continuous,17 the influx of immune cells to the spinal tissue is unlikely to have preceded TB infection. An influx of HIV-infected immune cells and free virions would explain the evidence of a “founder effect” that we observed through phylogenetic reconstruction. Furthermore, the close contact between HIV-infected lymphocytes and TB-infected macrophages may facilitate enhanced HIV replication or even cell-to-cell HIV spread.31 Despite high tissue viral loads, intracompartment diversity was low, consistent with the hypothesis of clonal expansion of HIV variants from a founder virus that migrated into the spinal TB granuloma. Furthermore, we showed greater viral diversity and evolutionary distance between plasma-derived and tissue-derived viruses, particularly when comparing adults with children; this is a reflection perhaps of the immature immune systems of children or more recent HIV acquisition where lower diversity is a characteristic of recent infection and early viral kinetics.
The immune response and physiology of children differ significantly from adults; however, our findings suggest that the phenomenon of viral sequestration within spinal TB granulomas may not be impacted by the age of the patient but may be influenced by the duration of infection. We do not know the precise timing of infection of either the adults or children but we speculate that adults may on average have been infected longer considering that disease progression is accelerated in children compared with adults32 and the advanced clinical disease (low CD4 and high VL) among adults in this cohort.
HIV population dynamics follow a strong temporal structure where sequences from samples taken from an individual at the same time point cluster together.12 It may be argued that a similar phenomenon is reflected in our findings of monophyletic clustering. However, the tree topologies and longer branch lengths we observed may equally be indicative of the impact of complex phylodynamic determinants such as immune selection pressure and site-specific population dynamics between latent/reactivated and productive viremia. Moreover, we demonstrated migration from the tissue to the plasma in two of six patients, both children (<5 years of age), suggesting that the contribution to the systemic viral population from the tissue may be limited. In contrast, Collins et al.15 noted frequent, unidirectional migration events from pleura to blood representative of a direct mechanism for greater systemic HIV heterogeneity.
The role of the V3 loop in viral tropism, cytopathicity, replication efficiency, and fusion is known.28 CCR5 exploiting variants predominate, even in the context of Mycobacteria coinfections.33 Our findings established CCR5-utilizing variants in both plasma and tissue in all but two sequences; this is consistent with previous studies of HIV-1 subtype C infections showing minimal corrector switch from CCR5 to CXCR4 even in the context of late stage AIDS and coinfections such as pulmonary TB.28 Phenotypic studies may be warranted since genotype does not always accurately predict coreceptor utilization and subtle differences in usage of CD4 and coreceptors have been noted with CSF-derived and other viral isolates.34,35
Given the prominent role of macrophages in the anti-TB immune response we explored their composition in the granulomas and found evidence of HIV-infected macrophages. In this study, we present electron micrographs of both HIV and TB within membrane-bound intracellular compartments. To the best of our knowledge, this is the first reported evidence of viral particles in endosomal compartments in spinal TB/HIV-coinfected tissue. We further demonstrated an association between viral diversity and macrophage infiltration of spinal granulomas suggesting that macrophages may be a possible cellular target of HIV infection in granulomas. This hypothesis is plausible for four reasons: (1) macrophages show susceptibility to HIV infection, (2) infiltration of differentiated immune cells to the site of infection, (3) long-lived archived viruses demonstrated within macrophages, and (4) SIV models showing HIV infection of monocytes and macrophages. In the first instance, susceptibility to HIV infection is increased in differentiated macrophages, which are abundant in TB granulomas, compared to undifferentiated monocyte-derived macrophages (MDM).36 Second, the influx of activated immune cells including differentiated macrophages to the site of infection regardless of HIV status and the concomitant HIV-related T cell depletion19 mean that an environment permissive to exploiting macrophages may be created in spinal TB granulomas. However, there remains controversy around this paradigm with conflicting reports showing that CD4 depletion either has an impact37 or has no impact38 on HIV infection of myeloid cells. Third, long-lived HIV lineages have previously been correlated with macrophage-targeting HIV variants in the CNS.7,35 Finally, it is noteworthy that myeloid cells (macrophages, monocytes, and dendritic cells) have been shown to become infected with HIV following phagocytosis of infected T cells in the SIV model of infection.38
A limitation of our study is the small sample size and the limited number of clones analyzed per compartment. The challenges in amplifying the HIV genome from tissue restricted our use of limiting dilution techniques such as single genome amplification, which have been shown to mitigate the problem of variant resampling within quasispecies.39–41 However, it has also been demonstrated that the problems of variant resampling, population structure differences, and sequencing bias can be overcome or reduced if a large enough number of clones is analyzed.42 Although we analyzed only an average of seven clones per compartment in our study, we employed rigorous quality checks at both the laboratory and analysis stages of our study to ensure that we did not confound the interpretation of viral population diversity. In this study, we did not explore the contribution of the CSF-derived variants to the viral population within these granulomas. In our study and previous clinical case reports it was shown that Mtb infection of the spine is extradural both in HIV-infected and HIV-uninfected patients43 and penetration of the meninges into the spinal cord is rare,44 thus maintaining the integrity of the blood–brain barrier. The possibility of viral trafficking between the granuloma and the CSF site cannot be ruled out, and this aspect needs to be explored in future investigations. Investigating the spinal granuloma as a site of HIV divergence also requires investigation of viral divergence in the context of ART, which we were unable to explore in the present study. Future prospective investigations will include HIV-infected patients on ART.
Conclusions
We show, in this small study, that spinal granulomas may form a site of independent viral evolution. If confirmed in larger studies, this may have implications for viral suppression and eradication strategies in sequestered sites based on whether antiretroviral therapy can penetrate these sites. We also show the possible involvement of macrophages in HIV replication within granulomas and raise the intriguing question of whether HIV exploits the survival strategies of TB to sequester within macrophages. Further studies to confirm these findings and to explore the accessibility of this anatomical niche by antiretroviral drugs and its implications for clinical disease progression, treatment, and eradication strategies are warranted.
Acknowledgments
The authors wish to thank all participants in this study and extend our appreciation to Dr. Ashwin Bramdev (Pathologist, PapLab) and Mrs. Keshni Hiramen (HIV Pathogenesis Programme) for their assistance. This work was funded by a Wellcome Trust Grant. T. Ndung'u is funded through the South African DST/NRF Research Chair in Systems Biology of HIV/AIDS, the Victor Daitz Chair in HIV/TB Research, and an International Early Career Scientist Award from the Howard Hughes Medical Institute.
All clonal sequences are available from the authors on request and are also available on GenBank (accession numbers: FJ394222–FJ394329).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Tuberculosis Fact sheet No 104. 2014. www.who.int/mediacentre/factsheets/fs104/en/ Accessed April7, 2014
- 2.Geldmacher C, Ngwenyama N, Schuetz A, et al. : Preferential infection and depletion of Mycobacterium tuberculosis-specific CD4 T cells after HIV-1 infection. J Exp Med 2010;207(13):2869–2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Svicher V, Ceccherini-Silberstein F, Antinori A, et al. : Understanding HIV compartments and reservoirs. Curr HIV/AIDS Rep 2014;11(2):186–194 [DOI] [PubMed] [Google Scholar]
- 4.Buzon MJ, Martin-Gayo E, Pereyra F, et al. : Long-term antiretroviral treatment initiated in primary HIV-1 infection affects the size, composition and decay kinetics of the reservoir of HIV-1 infected CD4 T cells. J Virol 2014;88:10056–10065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ndung'u T. and Weiss RA: On HIV diversity. AIDS 2012;26(10):1255–1260 [DOI] [PubMed] [Google Scholar]
- 6.Avettand-Fenoel V, Hocqueloux L, Muller-Trutwin M, et al. : Greater diversity of HIV DNA variants in the rectum compared to variants in the blood in patients without HAART. J Med Virol 2011;83(9):1499–1507 [DOI] [PubMed] [Google Scholar]
- 7.Schnell G, Price RW, Swanstrom R, and Spudich S: Compartmentalization and clonal amplification of HIV-1 variants in the cerebrospinal fluid during primary infection. J Virol 2010;84(5):2395–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fletcher CV, Staskus K, Wietgrefe SW, et al. : Persistent HIV-1 replication is associated with lower antiretroviral drug concentrations in lymphatic tissues. Proc Natl Acad Sci USA 2014;111(6):2307–2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghosn J, Viard JP, Katlama C, et al. : Evidence of genotypic resistance diversity of archived and circulating viral strains in blood and semen of pre-treated HIV-infected men. AIDS 2004;18(3):447–457 [DOI] [PubMed] [Google Scholar]
- 10.Choudhury B, Pillay D, Taylor S, and Cane PA: Analysis of HIV-1 variation in blood and semen during treatment and treatment interruption. J Med Virol 2002;68(4):467–472 [DOI] [PubMed] [Google Scholar]
- 11.Ritola K, Robertson K, Fiscus SA, et al. : Increased human immunodeficiency virus type 1 (HIV-1) env compartmentalization in the presence of HIV-1-associated dementia. J Virol 2005;79(16):10830–10834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salemi M: The intra-host evolutionary and population dynamics of human immunodeficiency virus type 1: A phylogenetic perspective. Infect Dis Rep 2013;5(Suppl 1):e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanjuan R, Codoner FM, Moya A, and Elena SF: Natural selection and the organ-specific differentiation of HIV-1 V3 hypervariable region. Evolution 2004;58(6):1185–1194 [DOI] [PubMed] [Google Scholar]
- 14.Marras D, Bruggeman LA, Gao F, et al. : Replication and compartmentalization of HIV-1 in kidney epithelium of patients with HIV-associated nephropathy. Nat Med 2002;8(5):522–526 [DOI] [PubMed] [Google Scholar]
- 15.Collins KR, Quinones-Mateu ME, Wu M, et al. : Human immunodeficiency virus type 1 (HIV-1) quasispecies at the sites of Mycobacterium tuberculosis infection contribute to systemic HIV-1 heterogeneity. J Virol 2002;76(4):1697–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Biru T, Lennemann T, Sturmer M, et al. : Human immunodeficiency virus type-1 group M quasispecies evolution: Diversity and divergence in patients co-infected with active tuberculosis. Med Microbiol Immunol 2010;199(4):323–332 [DOI] [PubMed] [Google Scholar]
- 17.Heath L, Fox A, McClure J, et al. : Evidence for limited genetic compartmentalization of HIV-1 between lung and blood. PLoS One 2009;4(9):e6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaufmann SH: Protection against tuberculosis: Cytokines, T cells, and macrophages. Ann Rheum Dis 2002;61(Suppl 2):ii54–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Danaviah S, Sacks JA, Kumar KP, et al. : Immunohistological characterization of spinal TB granulomas from HIV-negative and -positive patients. Tuberculosis (Edinb) 2013;93(4):432–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Govender S, Annamalai K, Kumar KP, and Govender UG: Spinal tuberculosis in HIV positive and negative patients: Immunological response and clinical outcome. Int Orthop 2000;24(3):163–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gordon M, De Oliveira T, Bishop K, et al. : Molecular characteristics of human immunodeficiency virus type 1 subtype C viruses from KwaZulu-Natal, South Africa: Implications for vaccine and antiretroviral control strategies. J Virol 2003;77(4):2587–2599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson JD, Higgins DG, and Gibson TJ: CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 1994;22(22):4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stamatakis A: Using RAxML to Infer Phylogenies. Curr Protoc Bioinformatics 2015;51. [DOI] [PubMed] [Google Scholar]
- 24.Ronquist F, Teslenko M, van der Mark P, et al. : MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 2012;61(3):539–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Posada D. and Crandall KA: MODELTEST: Testing the model of DNA substitution. Bioinformatics 1998;14(9):817–818 [DOI] [PubMed] [Google Scholar]
- 26.Pond SL, Frost SD, and Muse SV: HyPhy: Hypothesis testing using phylogenies. Bioinformatics 2005;21(5):676–679 [DOI] [PubMed] [Google Scholar]
- 27.Thielen A, Lengauer T, Swenson LC, et al. : Mutations in gp41 are correlated with coreceptor tropism but do not improve prediction methods substantially. Antivir Ther 2011;16(3):319–328 [DOI] [PubMed] [Google Scholar]
- 28.Ndung'u T, Sepako E, McLane MF, et al. : HIV-1 subtype C in vitro growth and coreceptor utilization. Virology 2006;347(2):247–260 [DOI] [PubMed] [Google Scholar]
- 29.Mlcochova P, Watters SA, Towers GJ, et al. : Vpx complementation of 'non-macrophage tropic' R5 viruses reveals robust entry of infectious HIV-1 cores into macrophages. Retrovirology 2014;11:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zarate S, Pond SL, Shapshak P, and Frost SD: Comparative study of methods for detecting sequence compartmentalization in human immunodeficiency virus type 1. J Virol 2007;81(12):6643–6651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sigal A. and Baltimore D: As good as it gets? The problem of HIV persistence despite antiretroviral drugs. Cell Host Microbe 2012;12(2):132–138 [DOI] [PubMed] [Google Scholar]
- 32.Mphatswe W, Blanckenberg N, Tudor-Williams G, et al. : High frequency of rapid immunological progression in African infants infected in the era of perinatal HIV prophylaxis. AIDS 2007;21(10):1253–1261 [DOI] [PubMed] [Google Scholar]
- 33.Santucci MB, Bocchino M, Garg SK, et al. : Expansion of CCR5+ CD4+ T-lymphocytes in the course of active pulmonary tuberculosis. Eur Respir J 2004;24(4):638–643 [DOI] [PubMed] [Google Scholar]
- 34.Aasa-Chapman MM, Aubin K, Williams I, and McKnight A: Primary CCR5 only using HIV-1 isolates does not accurately represent the in vivo replicating quasi-species. Virology 2006;351(2):489–496 [DOI] [PubMed] [Google Scholar]
- 35.Schnell G, Joseph S, Spudich S, et al. : HIV-1 replication in the central nervous system occurs in two distinct cell types. PLoS Pathog 2011;7(10):e1002286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aljawai Y, Richards MH, Seaton MS, et al. : Beta-catenin/TCF-4 signaling regulates susceptibility of macrophages and resistance of monocytes to HIV-1 productive infection. Curr HIV Res 2014;12(3):164–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ortiz AM, Klatt NR, Li B, et al. : Depletion of CD4(+) T cells abrogates post-peak decline of viremia in SIV-infected rhesus macaques. J Clin Invest 2011;121(11):4433–4445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Calantone N, Wu F, Klase Z, et al. : Tissue myeloid cells in SIV-infected primates acquire viral DNA through phagocytosis of infected T cells. Immunity 2014;41(3):493–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu SL, Rodrigo AG, Shankarappa R, et al. : HIV quasispecies and resampling. Science 1996;273(5274):415–416 [DOI] [PubMed] [Google Scholar]
- 40.Simmonds P: Variation in HIV virus load of individuals at different stages in infection: Possible relationship with risk of transmission. AIDS 1990;4(Suppl 1):S77–83 [PubMed] [Google Scholar]
- 41.Edmonson PF. and Mullins JI: Efficient amplification of HIV half-genomes from tissue DNA. Nucleic Acids Res 1992;20(18):4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jordan MR, Kearney M, Palmer S, et al. : Comparison of standard PCR/cloning to single genome sequencing for analysis of HIV-1 populations. J Virol Methods 2010;168(1–2):114–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Govender S, Parbhoo AH, Kumar KP, and Annamalai K: Anterior spinal decompression in HIV-positive patients with tuberculosis. A prospective study. J Bone Joint Surg Br 2001;83(6):864–867 [DOI] [PubMed] [Google Scholar]
- 44.Sharma SK. and Mohan A: Extrapulmonary tuberculosis. Indian J Med Res 2004;120(4):316–353 [PubMed] [Google Scholar]