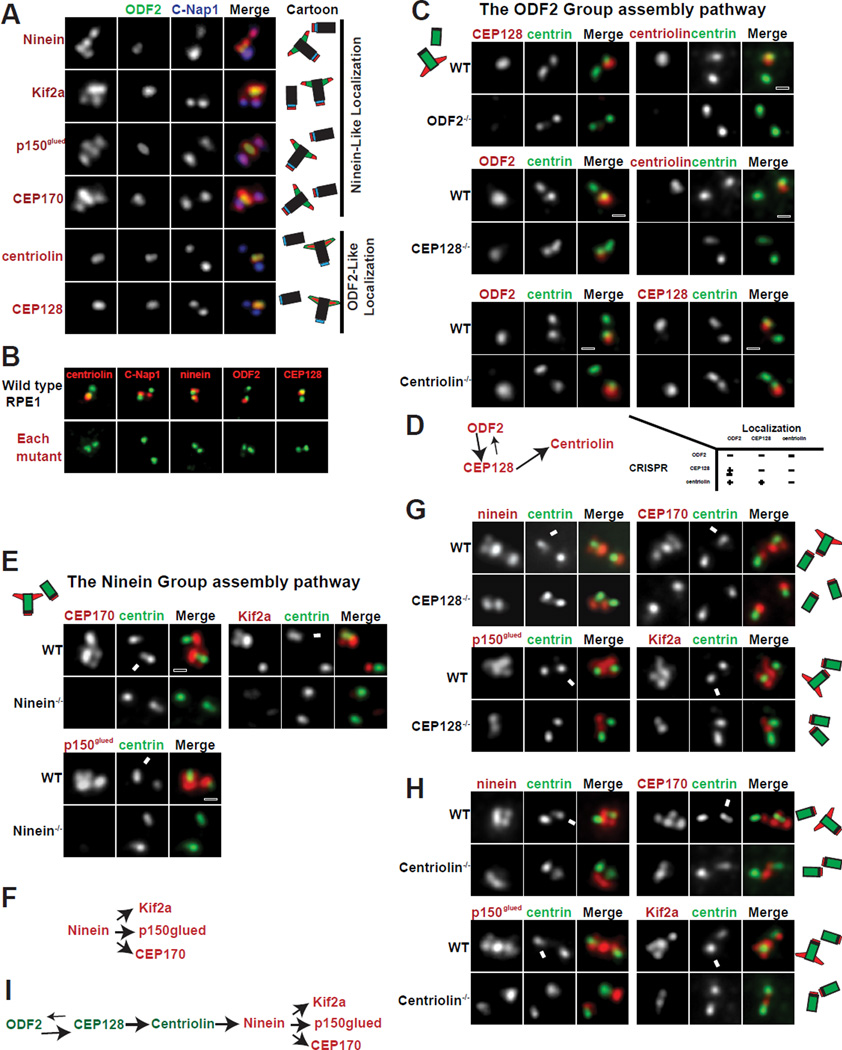
Figure 1. The assembly hierarchy of subdistal appendage components.
(A) The localization patterns of sDAP-associated proteins. Localization of respective proteins (red) was shown in relation to centrosome markers for proximal ends (C-Nap1 in blue) and subdistal appendages (ODF2 in green). Results are summarized in diagram and shown in respective colors.
(B) CRISPR-mediated knockout cell lines for each sDAP component (red). Mutant RPE1 cell lines were stained with indicated antibodies (red). Proteins targeted by CRISPR are shown in red along with centrosome markers in green.
(C), Wild-type (WT) or clonal RPE1 cells lines knocked out (KO) of indicated proteins by CRISPR are stained with indicated antibodies. Loss of ODF2 disrupts localization of CEP128 and centriolin at centrioles. Loss of CEP128 disrupts localization of centriolin from centrioles and reduces ODF2 levels at centrioles. Loss of centriolin has no effect on the localization of CEP128 nor ODF2.
(D) A schematic of the assembly hierarchy of the ODF2 group members.
(E) Loss of ninein impairs the localization of CEP170, Kif2a and p150glued to the centrosome.
(F) A schematic of the assembly hierarchy of the Ninein Group.
(G and H) The centrioles of wild-type RPE1 cells have 4 foci positive for each of the Ninein Group members. In CEP128 depleted or centriolin-depleted cells, the foci at the sDAP disappear, leaving only two foci. Arrows indicate the mature mother centrosome. (I) A schematic of the assembly pathway at the sDAP including both ODF2 Group and Ninein Group members.
See also Figure S1