Abstract
Continuum solvation representations based on the Poisson-Boltzmann equation have become widely accepted in biomolecular applications after years of basic research and development. Since analytical solution of the differential equation can be achieved only in a few specific cases with simple solute geometry, only numerical solution is possible for biomolecular applications. However, it is conceptually difficult to assign solvation forces in the numerical methods, limiting their applications into direct simulations of energy minimization and molecular dynamics. In this study a dielectric pressure formulation was derived from the general Maxwell stress tensor for continuum solvation of biomolecules modeled with the widely used abrupt-transitioned dielectrics. A charge-central strategy was then proposed to improve the numerical behavior of the computed pressure. An interesting observation is the highly similar charge-central formulations between the smooth-transition dielectric and the abrupt-transition dielectrics models utilized in the biomolecular solvation treatments. The connections of the new formulation with both the Davis-McCammon and Gilson et al. approaches were further presented after applying the normal field approximation. The consistency was verified with the numerical tests on a realistic biomolecule. The numerical experiments on the tested biomolecule further indicate that the charge-central strategy combined with the normal field approximation not only improves the accuracy of the dielectric boundary force but also reduces its grid dependence for biomolecular applications.
Introduction
Atomistic simulations have become an important tool for studying the structure, dynamics, and function of biomolecules. Since most particles in atomistic simulations are to represent water molecules solvating the target biomolecules, implicit treatment of water molecules allows greatly increased simulation efficiency. Indeed, implicit solvation treatments, or implicit solvents, offer a unique opportunity for more efficient simulations without the loss of atomic-level resolution for biomolecules. With constant community-wide development, implicit solvent models have emerged as simple and accurate alternatives for solvation interactions.1-14 The Poisson-Boltzmann implicit solvents, in particular, which are based on the Poisson-Boltzmann equation (PBE), have become widely accepted in biomolecular applications after over 20 years of basic research and development in the community.1-14 Since analytical solution of the PBE can be achieved only in a few specific cases with simple solute geometry, only numerical solution of the PBE is possible for biomolecular applications. Among the numerical solution methods for PBE, finite difference methods (FDM),15-32 finite element methods (FEM)33-39 and boundary element methods (BEM)40-54 are mostly used.
However, it is conceptually difficult to assign solvation forces in the numerical PBE methods, limiting their applications into direct simulations of molecular dynamics.25,41,55-61 Along with other limitations, the PBE is mostly solved for practical applications involving static conformations. To extend the application of the PBE to molecular dynamics and energy minimization, robust and efficient methodologies to compute solvation forces must be developed. It has been shown that solvation forces can be divided into two components: reaction field forces and dielectric pressure or boundary forces (DBF).25,41,55-60 The reaction field forces act upon charges so they can be applied directly to atoms. The DBF’s act upon the solute/solvent interface so they need to be further distributed to atoms in molecular dynamics. If electrolyte ions are present, there is a third component of solvation forces called the excess osmotic pressure, applied to the Stern layer. However, it is often not considered because of their much smaller magnitude. All these solvation forces are then added to the usual molecular mechanics force terms for molecular dynamics simulations. Apparently, energy conservation is very important to stable molecular dynamics, which relies on accurate force calculations. Thus computation of reaction field forces and DBF is key to the success of numerical PBE methods in molecular dynamics. Reaction field forces are straightforward to compute after the PBE is solved, but computation of dielectric boundary forces remains as a challenge.25,41,55-60
Much community-wide efforts have been devoted to the calculation of the DBF.25,55-57,62 The brute-force “virtual work” method is apparently the most definitive.55 However, use of the method requires at least four full numerical calculations to calculate each force vector. Apparently, this is only realistic for molecules treated as rigid bodies.
Davis and McCammon proposed a DBF formulation by examining the integration of the Maxwell stress tensor, the DBF surface density was shown as16,29,53
| (1) |
where εo and εi are the dielectric constants of the solvent and the solute, respectively, and Eo and Ei are the values of the electric field evaluated on the solvent and solute sides of the molecular surface, respectively, is the normal direction of the surface from the solute towards the solvent. Integration of this quantity over the surface yields the total dielectric boundary force for the molecule.
Che et al. revisited the DBF calculation through a variational strategy in the classical two-dielectric model.62 Given the assumption that the normal surface field contributes predominantly to DBF, they showed that the DBF can be formulated as
| (2) |
where ε∇ϕ represents the continuous normal dielectric displacement vector on the solute/solvent dielectric interface. This formulation was later updated by Li et al. in their second paper on deriving the DBF from the variation of the electrostatic free energy with respect to the location change of the dielectric boundary to63
| (3) |
The BEM is another promising approach to incorporate the continuum electrostatics into molecular mechanics simulations.41,44,64-68 The DBF calculation in the BEM using a polarization charge method was first described by Zauhar,56 who showed that the DBF can be calculated as
| (4) |
where σ is the surface polarization charge density. This expression was derived from eqn (1). The use of surface polarization charge density makes it straightforward in the BEM, where the Poisson’s equation can be solved through the iteration of the surface polarization charge density. Cortis et al. also tried to compute the DBF via the Maxwell stress tensor for their FEM, leading to the same formulation as that of Zauhar.34
Gilson et al. presented a variational approach for the DBF,57 and it was further tailored into a numerical algorithm for the FDM. Their expression for the DBF can be expressed as
| (5) |
where ε is the dielectric constant and E is the electric field. Note that the direction of the DBF in eqn (5) is consistent with that in eqn (1) and in its equivalent formulations because the gradient of dielectric transition is in the normal direction. Im et al. proposed a method equivalent to eqn (5) as25
| (6) |
where ϕ is the total electrostatic potential, r represents the atomic coordinates. Apparently both eqn (5) and (6) require smoothly varying dielectric models since ∇ε has to be finite, i.e. ε has to be designed to change from εi to εo sufficiently smoothly for stable numerical performance.69 This would exclude the classical two-dielectric model where ∇ε is infinite. Even if the harmonic average is used at the solute/solvent dielectric interface, direct numerical calculation of ∇ε is still unstable in molecular dynamics. This leads to large and unstable DBF that does not satisfy the “virtual work” principle. Numerical approximation of ∇ε may alleviate the problem, but instability cannot be completely eliminated in molecular dynamics.57,60
Very recently we proposed a new DBF formulation for FDM based on the concept of boundary polarization charge as61
| (7) |
where ρpol is the boundary polarization charge density, D is the electric displacement vector, and Dn is the normal component of D. This “charge-based” method also works for dielectric models that are smooth-transitioned.
In this study, we intended to address the more challenging theoretical issue in modeling the DBF when the commonly used abrupt-transitioned dielectric framework is adopted in modeling biomolecular electrostatic solvation. In the rest of the manuscript, a DBF formulation based on the general Maxwell stress tensor is first proposed. This is followed with discussions of its relationship with some of the existing DBF formulations as described in the Introduction. The new formulation was verified by numerical implementations for the FDM solutions of a realistic model peptide. Several numerical strategies, including the charge-central strategy, the normal field approximation, and the use of singularity removal in the solution of the potential, are incorporated to improve its numerical performance.
Methods
1. Derivation of DBF based on the Maxwell stress tensor
In our previous work, we proposed a new DBF formulation based on the differential approach of the Maxwell stress tensor for the smooth-transition dielectric treatment at the solute/solvent interface.61 However, the differential approach is limited in that it cannot be applied to the abrupt-transition dielectric treatment for which it is often more straightforward to enforce the classical dielectric interface jump conditions.
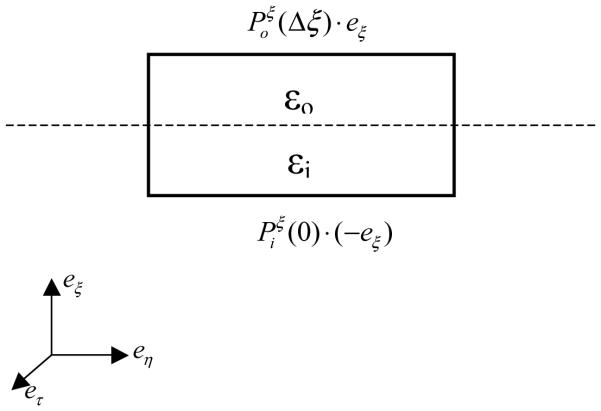
An alternative strategy is the integral approach, where a thin volume element is chosen on the interface as shown in Figure 1. The total force acting on the volume element can be computed by the surface integration of the Maxwell stress tensor (P) on its six faces. In the normal direction, the force density can be written as
| (8) |
Here eξ is the outward-directed normal unit vector of the molecular surface, and and are, respectively, the stress tensors on the surfaces parallel to dielectric interface inside and outside of the solute (Figure 1). Based on the definition of stress tensor, we have
| (9) |
Figure 1.
A volume element across the dielectric boundary. Dashline: dielectric boundary; i: solute region; o: solvent region. The sign convention follows the local coordinate system at the lower left corner.
Similarly can be computed as
| (10) |
Here eη and eτ are the two orthogonal tangential unit vector of the molecular surface.
Given the jump conditions at the dielectric interface
| (11) |
eqn (9), and eqn (10), the normal force density (eqn (8)) can be written in terms of field components as
| (12) |
Since the thickness of the small volume is a higher-order infinitesimal, the tangential components can be neglected. Thus the dielectric boundary force density only exists in the normal direction of the surface, and we have
| (13) |
2. Charge-based formulation
We can also develop a charge-based strategy for the abrupt-transition dielectric treatment, similar to our prior development for the smooth-transition dielectric treatment.61 In the following, we first transform eqn (13) in terms of the surface charge density.
We first note that in the boundary region the Gauss’ law gives
| (14) |
where σpol is the surface charge density. The tangential component of E is continuous across the dielectric interface, so that the differential form of eqn (14) can be written as
| (15) |
Substituting jump condition εoEoξ = εiEiξ into eqn (15), we have
| (16) |
Or equivalently .
Given this preparation we now return to eqn (13). First it is transformed into field components as
| (17) |
Utilization of boundary conditions eqn (11) gives
| (18) |
Given and εoEoξ = εiEiξ, eqn (18) can then be reformulated as
| (19) |
or in the equivalent and cleaner form if electric displacement is used
| (20) |
where Do and Di are the corresponding electric displacements of the solvent and solute sides, respectively. Note that eqn (20) is different from the Zauhar’s formulation of eqn (4),53 which also relies on the knowledge of surface charge density: eqn (20) can be translated into a sum of surface charges that is more suitable for the FDM.
An interesting observation is the similarity of eqn (20) and the charge-based approach for the smooth-transition dielectric treatment61
| (21) |
Of course, volume density and integration should be used in the smooth-transition dielectric model because there is no longer a sharp interface between the solvent and solute. However, the basic operation is still the same where the polarization charges and electric displacements are needed in the region of non-uniform dielectric constant.
3. Relations to other DBF expressions
It can be shown that eqn (13) is consistent with other expressions for DBF for the abrupt-transition dielectric treatment. For example, eqn (13) can be transformed into the formulation (eqn (1)) by Davis and McCammon55 given the jump conditions in eqn (11) as shown in Appendix. Of course, it is also consistent with eqn (3) as proposed by Zauhar, which is derived from eqn (1).56 Apparently, the formulation is different from that of the Gilson et al.,57 which was derived for the smooth-transition dielectric treatment. However, it is interesting to note that the two charge-based formulations are remarkably similar whether the abrupt transition or smooth transition is used in the interface dielectric treatment as discussed above.
4. Numerical algorithms
Numerical implementation of eqn (13) requires integration over the molecular surface. Here we utilized a fast FDM approximation to the molecular surface to conduct the surface integration.70 Given that a molecular surface is defined by a set of solute atomic spheres and a solvent probe sphere, we can always compute the intersection points between the molecular surface and any intersecting FDM grid edge with a numerical procedure.71 Indeed, these intersection points form a very decent representation of the brute-force molecular surface for visualization proposes. In the fast FDM approximation of molecular surface, a molecular surface element (s) can be approximated as follows.70
| (22) |
where d is the x/y/z distance between the center of an edge intersected by the molecular surface and the atom/probe center if the edge is an x/y/z edge, respectively, r is the distance between the center of the edge and the atom/probe center, R is the radius of the atom/probe. As shown in a separate publication, the fast FDM approximation converges to the analytical molecular surface for a large set of diversified molecules, including both proteins and nucleic acids.70 Thus, the DBF can be computed as
| (23) |
where n is the number of intersection points between the grid edges and the analytical molecular surface.
To discretize the charge-based method, we utilized the following assumption. In the FDM, the polarization charges can be regarded as being located at the boundary grid points.28 Given this approximation and eqn (20), the DBF surface integration can be written as
| (24) |
where n is the number of the polarization charges, is the value of the polarization charge on the ith boundary grid point.
It should also be pointed out that there is naturally a second way to discretize eqn (20): we can simply use (15) to compute the surface polarization charge at the intersection points. Of course, such a discretized charge-based method is numerically identical to eqn (23). Thus we will not analyze the second strategy below. Finally the surface polarization charges are calculated using the Gauss’ law and the grid potential map obtained from the FDM.28 The boundary grid polarization charges are projected on the molecular surface according to the procedure described by Rocchia et al.28
4. Normal field approximation
Conductor approximation
Due to the typical high value of dielectric constant of water versus that of the solute in molecular mechanics force fields, the tangential surface field is often extremely small when compared with the normal surface field. This is because water can almost be approximated as a conductor if the dielectric constant of the molecular interior is set to 1 as in typical molecular mechanics simulations, i.e. the ratio of exterior and interior dielectric constants is ~O(102). This is the underlining assumption in the COSMO model for quantum mechanical calculations.72 In this setting, the argument of electrostatic equilibrium for the conductor/vacuum interface can be applied to the water/vacuum interface without much error: the surface charge distribution can only reach electrostatic equilibrium when the surface tangential field is zero.
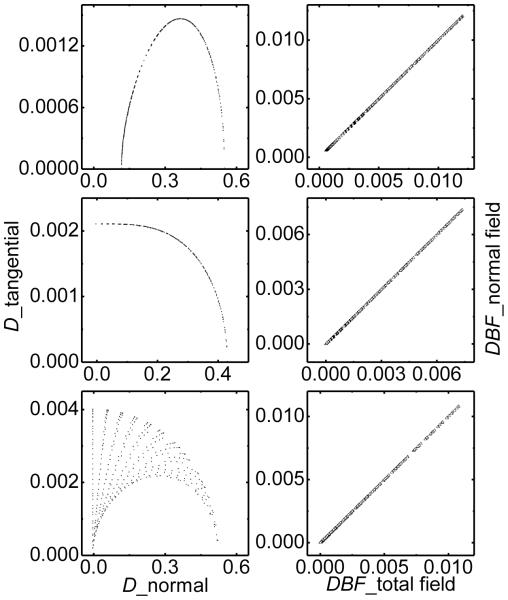
We used a well-studied testing system, a single dielectric sphere imbedded with point charges, to study the validity of the normal field approximation. The radius of the sphere R is 2.0 Å, about the size of a united carbon atom, centered at (0, 0, 0). The dielectric constant inside is set as 1.0, the dielectric constant outside is set as 80.0. Three typical situations are used: (a) charged system with a unit positive charge positioned 0.5 Å away from the center; (b) dipolar system with two unit charges located at (−0.5, 0, 0) and (0.5, 0, 0), respectively. (c) quadrupolar system with four unit charges located at (−0.5, 0.5, 0), (−0.5, −0.5, 0), (0.5, −0.5, 0) and (0.5, 0.5, 0). As shown in Figure 2, the tangential D is much smaller than the corresponding normal D (less than 1%) for all the three testing systems. The DBF correlations between the normal field approximation and total field method are quite good.
Figure 2.
Normal field approximation in the single-charged arrangement (top), dipolar-charged arrangement (middle), and quadrupolar-charged arrangement (bottom), respectively. Left: the correlation of the normal electrical displacement and the tangential electrical displacement. Right: The DBF correlation between and total field and normal field approximation. The force and fields are in electron-Ångstrom unit (e2/Å2) for the model system calculations.
Thus, use of the normal component of the field alone is a reasonable choice given the more difficult task of interpreting the total field, or interpreting the extremely small tangential field. As will be shown below, this can be utilized to improve the convergence of numerical DBF, particularly at coarser grid spacings.
Specifically the field-based method, eqn (23), can be approximated, under the normal field approximation, as
| (25) |
Similarly the charge-based method, eqn (24), can be approximated as
| (26) |
Consequence of the normal field approximation
Eqn (22) offers second-order accuracy in the calculation of molecular surface areas.70 If we use a first-order approximation of eqn (22) in eqn (25), i.e.,
| (27) |
where is used, d' is the x/y/z distance between the intersection point and the center of the atom/probe, r is the distance between the intersection point and the center of the atom/probe, then eqn (25) becomes
| (28) |
To discretize the formulation of Gilson et al. (see eqn (5)) we need the following relation given the harmonic averaging treatment at the fractional x-edge57
| (29) |
Eqn (5) can then be rewritten as
| (30) |
where Ex is the electric field in the x direction at the intersection point. Since Dx is the x component of Diξ (Diξ cos θ = Dx), eqn (28) is thus equivalent to eqn (30).
In summary eqn (13) can be cast into a discretized form (eqn (28)) that is mathematically consistent with the discretized form (eqn (30)) of the Gilson et al. approach57 given that (a) the harmonic averaging is used and (b) the normal field approximation is used. Of course, the discretized form of eqn (28) has an advantage of without the division by cos θ , which is the source of numerical instability in eqn (30). Thus, we can also achieve the goal of reducing the numerical instability in the DBF calculation by resorting to a different strategy as given by eqn (28).
6. Computational details
Surface field calculation
We utilized the one-side first-order least-square interpolation method to obtain the surface potential and field at an arbitrary position (x0, y0, z0).73 Briefly a potential function of the form
| (31) |
is fitted using the potentials of Nm (≥ 4) nearest grid points in the same dielectric. The coefficients ai, (i = 0,1,2,3) are determined to minimize
| (32) |
so that the potential and derivative of potential at position (x0, y0, z0) is given by the following relation:
| (33) |
Charge singularity removal
To improve the quality of surface field calculation, we used the singularity-free numerical solution of the PB equation. Briefly, the PB equation can be reformulated into two separate equations for two different potentials in two different regions that can be solved simultaneously:74
| (34) |
The reaction field potential (ϕRF) is solved in the solute region (Ωi) and the total potential (ϕ = ϕC + ϕRF) is solved in the solvent region (Ωo). Here ϕC is the Coulombic potential, satisfying εi∇2ϕC = −ρ and f(ϕ) is the Boltzmann salt term and λ is the Stern layer masking function. The interface conditions across the interface Γ are74
| (35) |
Interface treatments
In biomolecular calculations the dielectric distribution often adopts a piece-wise constant model. In such a model, the dielectric constant at a midpoint apparently should be assigned to the dielectric constant in this region where the two neighbor grid points belong. However, when the two neighbor grid points belong to different dielectric regions, its dielectric constant is nontrivial to assign, because the dielectric constant is discontinuous across the interface. One simple treatment is the use of harmonic average (HA) of the two dielectric constants at the interface midpoints.75 This strategy has been shown to improve the convergence of reaction field energies with respect to the grid spacing.75
Data Analysis
An issue important for stable dynamics simulation is the numerical uncertainty of solvation forces when the finite-difference grid is randomly positioned. Sensitivity of grid positions with respect to the solute molecule has been a particularly annoying limitation in current finite-difference PB methods. Here a total of 96 different finite-difference grid orientations were used to analyze the numerical uncertainty of the methods, i.e. the effect of relative location of finite-difference grids with respect to the interface and charge distribution. The dielectric interface between the solute and solvent regions was defined by the solvent-excluded molecular surface, obtained with a solvent probe radius 1.4 Å. The dielectric constant outside the solute is set as 1.0, the dielectric constant inside the solute is set as 80.0. The grid spacing ranges from 1/2 Å to 1/16 Å. The finite-difference convergence criterion was set to be 10−9. All molecular structures were first processed with Leap in AMBER 1276 and the modified Bondi radii were used except the radii of all hydrogen atoms were changed to 1.0 Å. All other parameters are set to be default as in the PBSA program of AMBER 12.27,60,76
Results and discussion
1. Consistency between field-based and charge-based strategies
First we compared the numerical performance of the field-based and charge-based algorithms. Here the numerical stability and convergence is demonstrated by the standard deviations, regression slopes, correlation coefficients (CC) and root mean squared (RMS) deviations (with respect to the forces at the finest grid spacing tested (1/16 Å)) of atomic DBF’s from 96 FDPB calculations with random orientations. The tested molecule is a helical structure with 281 atoms. As shown in Table 1, the total field strategy for atomic DBF’s performs poorly, especially at coarse grid spacings. For example, the slope at 1/2 Å is 1.464, which means an overall around 50% error. In contrast, the charge-based method shows much better consistency between the results at all grid spacings, as demonstrated by the fact that the slope is always very close to 1 and the RMS deviation is smaller than that of the field-based method. Furthermore, the fluctuations of the atomic forces by the charge-based method are also smaller, suggesting less significant grid dependence.
Table 1.
Convergence of atomic contact dielectric boundary (DB) forces and total electrostatic forces for the model helix by the field-based and charge-based methods. Total field at the dielectric boundary is used. The data at 1/16 Å is used as reference to compute the slope, CC, and rmsd for the data at 1/2, 1/4, and 1/8 Å. rmsd: root mean square deviation; σ: standard deviation; CC: correlation coefficient; NA: not available; unit of rmsd, σ and mean: kcal/(mol·Å).
| 1/h | DB force | Total force | |||||
|---|---|---|---|---|---|---|---|
| slope | CC | rmsd | σ | mean | σ | ||
| Field | 2 4 8 16 |
1.464 1.102 1.001 NA |
0.95217 0.99415 0.99984 NA |
1.152 0.273 0.028 NA |
0.249 0.050 0.010 0.003 |
146.877 39.309 6.698 0.955 |
12.335 2.563 0.450 0.292 |
| Charge | 2 4 8 16 |
0.999 1.008 1.004 NA |
0.99946 0.99988 0.99998 NA |
0.051 0.028 0.011 NA |
0.056 0.019 0.006 0.002 |
10.282 3.231 1.732 0.184 |
8.351 3.550 0.851 0.451 |
Convergence of the total electrostatic force (solvation electrostatic forces, including qE forces and DBF) to zero is also an important issue in MD simulations. It can be seen in Table 1, the standard deviations of total electrostatic forces are reduced and the mean total forces converge to zero when the grid spacing is reduced for both methods, indicating more accurate total forces at finer grid spacing. However, the mean total force by the field-based method at 1/2 Å is unacceptably large, 146.877 kcal/(mol· Å), although the field-based method converges very rapidly, for example, the mean total force is more than 150 times smaller when the grid spacing decreases from 1/2 Å to 1/16 Å. The mean total force by the charge-based method is more reasonable, but its fluctuation is on the same order as that by the field-based method.
2. Improvements with the normal field approximation
The results of the normal field approximation are shown in Table 2. Interestingly the atomic DBF’s obtained with the normal field approximation converge much better than those with the total field method. For the field-based method, both the regression slopes of field-based DBF’s and the mean total forces are improved at all grid spacings. For example, the slopes at 1/2 Å and 1/4 Å are 0.951 and 0.995, respectively, i.e., the overall errors are reduced by more than 10 times; the mean total force at 1/2 Å is reduced by nine times. The reduction in the mean total force can also be observed for the charge-based method at all grid spacings (see Table 2). In addition, the standard deviations of total forces become smaller for both methods after the normal field approximation is used, so the normal field approximation helps reduce grid dependence.
Table 2.
Convergence of atomic contact dielectric boundary (DB) forces and total electrostatic forces for the model helix by the field-based and charge-based methods. Normal field at the dielectric boundary is used. The data at 1/16 Å is used as reference to compute the slope, CC, and rmsd for the data at 1/2, 1/4, and 1/8 Å. Unit of rmsd, σ, and mean: kcal/(mol·Å).
| 1/h | DB force | Total force | |||||
|---|---|---|---|---|---|---|---|
| slope | CC | rmsd | σ | mean | σ | ||
| Field | 2 4 8 16 |
0.951 0.995 0.995 NA |
0.99940 0.99989 0.99998 NA |
0.096 0.024 0.013 NA |
0.057 0.019 0.006 0.002 |
16.145 8.959 4.857 0.365 |
3.610 0.836 1.020 0.061 |
| Charge | 2 4 8 16 |
0.992 1.006 1.003 NA |
0.99957 0.99990 0.99998 NA |
0.047 0.025 0.011 NA |
0.053 0.018 0.006 0.002 |
4.817 1.133 0.360 0.034 |
2.362 0.474 0.176 0.109 |
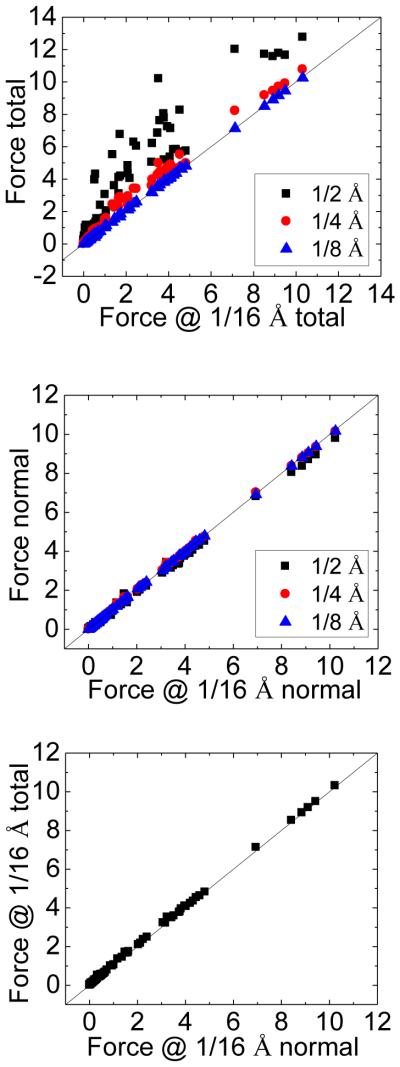
The benefit of the normal field approximation over the original method can also be shown in Figure 3, which plots the correlations between atomic DBF’s at the grid spacings of 1/2 Å, 1/4 Å, and 1/8 Å respectively, and those at the grid spacing of 1/16 Å. These plots are consistent with the correlation analysis presented in Table 2, indicating that the normal field approximation converges faster than the total field method. Finally the correlation between the total field method and the normal field approximation at the grid spacing of 1/16 Å is shown at the bottom panel of Figure 3, which verifies that the two methods finally converge to the same results as the grid spacing is reduced.
Figure 3.
Correlations between the contact dielectric boundary forces computed at the grid spacings of 1/2 Å, 1/4 Å, 1/8 Å, respectively, and those at the grid spacing of 1/16 Å for the model helix. Top: no approximation. Middle: normal field approximation. Bottom: no approximation at 1/16 Å. The correlation the between total field and normal field approximations at the grid spacing of 1/16 Å is shown at the bottom panel. Unit: kcal/(mol·Å).
In summary, the normal field approximation is preferable in computing the dielectric boundary force because it avoids interpolation of the tangential component of the field on the molecular interface, which is much smaller in magnitude than the normal component.
3. Connection and comparison with the Gilson et al. strategy
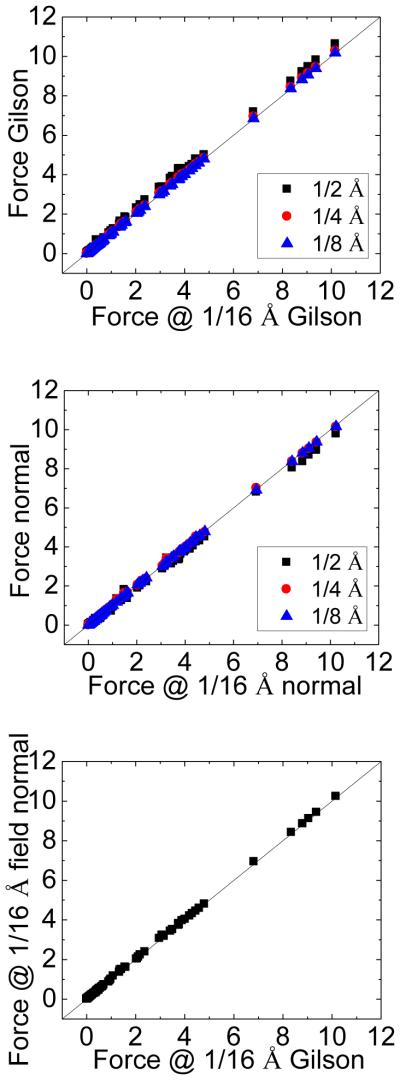
As discussed in the methods, the field-based method under the normal field approximation (eqn (25)) is mathematically consistent with the method by Gilson et al. if the HA dielectric treatment and the normal field approximation is used at the interface. Again we use the model helix as an example to demonstrate their consistency. The consistency between the two methods is shown in Figure 4, which plots the correlations between atomic DBF’s at 1/2 Å, 1/4 Å, 1/8 Å, respectively, and those at the grid spacing of 1/16 Å. It can be seen that the two methods have similar convergence performance at coarse grid spacings (see top panel and middle panel in Figure 4). The correlation between the normal field approximation and Gilson et al. at the grid spacing of 1/16 Å is also shown at the bottom panel of Figure 4, which verifies that the two methods are numerically consistent, in agreement with the mathematical analysis in Methods.
Figure 4.
Correlations between the contact dielectric boundary forces computed at the grid spacings of 1/2 Å, 1/4 Å and 1/8 Å respectively, and those at the grid spacing of 1/16 Å for the model helix. Top: Gilson et al. Middle: normal field approximation. Bottom: the correlation between Gilson et al. and normal field approximation at the grid spacing of 1/16 Å. Unit: kcal/(mol·Å).
Conclusion
We have derived a dielectric pressure formulation based on the general Maxwell stress tensor for continuum electrostatic solvation of biomolecules modeled with the abrupt-transitioned dielectrics framework. Several numerical strategies, including the charge-central strategy, the normal field approximation, and the use of singularity removal, are incorporated in its numerical implementation for the finite-difference numerical treatment of biomolecular solvation. An interesting observation is the highly similar charge-central formulations between the smooth-transition dielectric and the abrupt-transition dielectrics frameworks. Its connections with both Davis-McCammon and Gilson et al. formulations were also discussed. Interestingly, if normal field approximation is coupled with the harmonic average of interface dielectric constant, these methods are all mathematically consistent with each other, at least in the numerical implementation for the finite-difference treatment of solvation.
Our numerical tests show that the charge-based formulation offers much better consistency between the results at all grid spacings, as demonstrated by the fact that the slope is always very close to 1 and the RMS deviation is smaller than that of the field-based method. Furthermore, the fluctuations of the atomic forces by the charge-based method are also smaller, suggesting less significant grid dependence. The mean total force by the charge-based formulation is more reasonable, but its fluctuation is on the same order as that by the field-based method.
It is found that the numerical forces can be further improved when the normal field approximation is applied. This is the case for both field-based and charge-based formulations. The slopes and the standard deviations of atomic forces by both field-based and charge-based methods are improved with the normal field approximation. The results also indicate that the normal field approximation strategy converges faster than the total field method. The correlation between the total field method and the normal field approximation strategy at the grid spacing of 1/16 Å verifies that the two methods converge to the same results as the gird spacing is reduced. The convergence of total electrostatic forces is also analyzed. It is found the standard deviations of the total field method are about 4 – 10 times larger than those of the normal field approximation strategy.
It is instructive to discuss the future direction after establishing a physical rigorous formulation as presented here. Apparently, the numerical accuracy of both the linear system solver and the force interpolation method discussed here are still too low for MD simulations at the typical coarse grid spacing, i.e. 1/2 Å. To further improve the performance of both aspect of the numerical PB method, smooth molecular surface definitions must be explored so that a second order accurate numerical solver and surface field interpolation method can be used to improve the accuracy of the overall continuum solvation method for broader biomolecular applications.
Acknowledgements
We are grateful to research supports from NIGMS (R01GM093040 and R01GM79383).
Appendix.
Here we present a proof that the formulation of DBF (eqn (13))
is equivalent to Davis and Mccammon’s (eqn (1)). First eqn (13) is transformed into field components as
Utilization of boundary conditions εoEoξ = εiEiξ, Eoη = Eiη, and Eoτ = Eiτ gives
Further simplification gives
References
- (1).Davis MEME, McCammon JA. Chem. Rev. 1990;90:509. [Google Scholar]
- (2).Sharp KA. Curr. Opin. Struct. Biol. 1994;4:234. [Google Scholar]
- (3).Gilson MK. Curr. Opin. Struct. Biol. 1995;5:216. doi: 10.1016/0959-440x(95)80079-4. [DOI] [PubMed] [Google Scholar]
- (4).Honig B, Nicholls A. Sci. 1995;268:1144. doi: 10.1126/science.7761829. [DOI] [PubMed] [Google Scholar]
- (5).Roux B, Simonson T. Biophys. Chem. 1999;78:1. doi: 10.1016/s0301-4622(98)00226-9. [DOI] [PubMed] [Google Scholar]
- (6).Cramer CJ, Truhlar DG. Chem. Rev. 1999;99:2161. doi: 10.1021/cr960149m. [DOI] [PubMed] [Google Scholar]
- (7).Bashford D, Case DA. Annu. Rev. Phys. Chem. 2000;51:129. doi: 10.1146/annurev.physchem.51.1.129. [DOI] [PubMed] [Google Scholar]
- (8).Baker NA. Curr. Opin. Struct. Biol. 2005;15:137. doi: 10.1016/j.sbi.2005.02.001. [DOI] [PubMed] [Google Scholar]
- (9).Chen JH, Im WP, Brooks CL. J. Am. Chem. Soc. 2006;128:3728. doi: 10.1021/ja057216r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Feig M, Chocholousova J, Tanizaki S. Theor. Chem. Acc. 2006;116:194. [Google Scholar]
- (11).Im W, Chen JH, Brooks CL. Peptide Solvation and H-Bonds. Vol. 72. Elsevier Academic Press Inc; San Diego: 2006. p. 173. [Google Scholar]
- (12).Koehl P. Curr. Opin. Struct. Biol. 2006;16:142. doi: 10.1016/j.sbi.2006.03.001. [DOI] [PubMed] [Google Scholar]
- (13).Lu BZ, Zhou YC, Holst MJ, McCammon JA. Commun. Comput. Phys. 2008;3:973. [Google Scholar]
- (14).Wang J, Tan CH, Tan YH, Lu Q, Luo R. Communications in Computational Physics. 2008;3:1010. [Google Scholar]
- (15).Klapper I, Hagstrom R, Fine R, Sharp K, Honig B. Proteins. 1986;1:47. doi: 10.1002/prot.340010109. [DOI] [PubMed] [Google Scholar]
- (16).Gilson MK, Sharp KA, Honig BH. J. Comput. Chem. 1988;9:327. [Google Scholar]
- (17).Davis ME, McCammon JA. J. Comput. Chem. 1989;10:386. [Google Scholar]
- (18).Nicholls A, Honig B. J. Comput. Chem. 1991;12:435. [Google Scholar]
- (19).Luty BA, Davis ME, McCammon JA. J. Comput. Chem. 1992;13:1114. [Google Scholar]
- (20).Holst M, Kozack RE, Saied F, Subramaniam S. J. Biomol. Struct. Dyn. 1994;11:1437. doi: 10.1080/07391102.1994.10508078. [DOI] [PubMed] [Google Scholar]
- (21).Forsten KE, Kozack RE, Lauffenburger DA, Subramaniam S. J. Phys. Chem. 1994;98:5580. [Google Scholar]
- (22).Holst M, Saied F. J. Comput. Chem. 1993;14:105. [Google Scholar]
- (23).Madura JD, Briggs JM, Wade RC, Davis ME, Luty BA, Ilin A, Antosiewicz J, Gilson MK, Bagheri B, Scott LR, McCammon JA. Comput. Phys. Commun. 1995;91:57. [Google Scholar]
- (24).Bashford D. Lecture Notes in Computer Science. 1997;1343:233. [Google Scholar]
- (25).Im W, Beglov D, Roux B. Comput. Phys. Commun. 1998;111:59. [Google Scholar]
- (26).Rocchia W, Alexov E, Honig B. J. Phys. Chem. B. 2001;105:6507. [Google Scholar]
- (27).Luo R, David L, Gilson MK. J. Comput. Chem. 2002;23:1244. doi: 10.1002/jcc.10120. [DOI] [PubMed] [Google Scholar]
- (28).Rocchia W, Sridharan S, Nicholls A, Alexov E, Chiabrera A, Honig B. J. Comput. Chem. 2002;23:128. doi: 10.1002/jcc.1161. [DOI] [PubMed] [Google Scholar]
- (29).Wang J, Cai Q, Li ZL, Zhao HK, Luo R. Chem. Phys. Lett. 2009;468:112. doi: 10.1016/j.cplett.2008.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Cai Q, Wang J, Zhao HK, Luo R. J. Chem. Phys. 2009;130:145101. doi: 10.1063/1.3099708. [DOI] [PubMed] [Google Scholar]
- (31).Cai Q, Hsieh MJ, Wang J, Luo R. J. Chem. Theory Comput. 2010;6:203. doi: 10.1021/ct900381r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Wang J, Luo R. J. Comput. Chem. 2010;31:1689. doi: 10.1002/jcc.21456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).You TJ, Harvey SC. J. Comput. Chem. 1993;14:484. [Google Scholar]
- (34).Cortis CM, Friesner RA. J. Comput. Chem. 1997;18:1591. [Google Scholar]
- (35).Holst M, Baker N, Wang F. J. Comput. Chem. 2000;21:1319. [Google Scholar]
- (36).Baker N, Holst M, Wang F. J. Comput. Chem. 2000;21:1343. [Google Scholar]
- (37).Chen L, Holst MJ, Xu JC. SIAM J. Numer. Anal. 2007;45:2298. [Google Scholar]
- (38).Shestakov AI, Milovich JL, Noy A. J. Colloid Interface Sci. 2002;247:62. doi: 10.1006/jcis.2001.8033. [DOI] [PubMed] [Google Scholar]
- (39).Xie D, Zhou S. BIT Numerical Mathematics. 2007;47:853. [Google Scholar]
- (40).Miertus S, Scrocco E, Tomasi J. Chem. Phys. 1981;55:117. [Google Scholar]
- (41).Zauhar RJ, Morgan RS. J. Mol. Biol. 1985;186:815. doi: 10.1016/0022-2836(85)90399-7. [DOI] [PubMed] [Google Scholar]
- (42).Hoshi H, Sakurai M, Inoue Y, Chujo R. J. Chem. Phys. 1987;87:1107. [Google Scholar]
- (43).Zauhar RJ, Morgan RS. J. Comput. Chem. 1988;9:171. [Google Scholar]
- (44).Juffer AH, Botta EFF, Vankeulen BAM, Vanderploeg A, Berendsen HJC. J. Comput. Phys. 1991;97:144. [Google Scholar]
- (45).Rashin AA. J. Phys. Chem. 1990;94:1725. [Google Scholar]
- (46).Yoon BJ, Lenhoff AM. J. Comput. Chem. 1990;11:1080. [Google Scholar]
- (47).Zhou HX. Biophys. J. 1993;65:955. doi: 10.1016/S0006-3495(93)81094-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Bharadwaj R, Windemuth A, Sridharan S, Honig B, Nicholls A. J. Comput. Chem. 1995;16:898. [Google Scholar]
- (49).Purisima EO, Nilar SH. J. Comput. Chem. 1995;16:681. [Google Scholar]
- (50).Liang J, Subramaniam S. Biophys. J. 1997;73:1830. doi: 10.1016/S0006-3495(97)78213-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Vorobjev YN, Scheraga HA. J. Comput. Chem. 1997;18:569. [Google Scholar]
- (52).Totrov M, Abagyan R. Biopolymers. 2001;60:124. doi: 10.1002/1097-0282(2001)60:2<124::AID-BIP1008>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- (53).Boschitsch AH, Fenley MO, Zhou HX. J. Phys. Chem. B. 2002;106:2741. [Google Scholar]
- (54).Lu BZ, Cheng XL, Huang JF, McCammon JA. Proc. Natl. Acad. Sci. U. S. A. 2006;103:19314. doi: 10.1073/pnas.0605166103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Davis ME, McCammon JA. J. Comput. Chem. 1990;11:401. [Google Scholar]
- (56).Zauhar RJ. J. Comput. Chem. 1991;12:575. [Google Scholar]
- (57).Gilson MK, Davis ME, Luty BA, McCammon JA. J. Phys. Chem. 1993;97:3591. [Google Scholar]
- (58).Cortis CM, Friesner RA. J. Comput. Chem. 1997;18:1570. [Google Scholar]
- (59).Friedrichs M, Zhou RH, Edinger SR, Friesner RA. J. Phys. Chem. B. 1999;103:3057. [Google Scholar]
- (60).Lu Q, Luo R. Journal of Chemical Physics. 2003;119:11035. [Google Scholar]
- (61).Cai Q, Ye X, Wang J, Luo R. Chemical Physics Letters. 2011;514:368. doi: 10.1016/j.cplett.2011.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Che J, Dzubiella J, Li B, McCammon JA. J. Phys. Chem. B. 2008;112:3058. doi: 10.1021/jp7101012. [DOI] [PubMed] [Google Scholar]
- (63).Li B, Cheng X, Zhang Z. Siam Journal on Applied Mathematics. 2011;71:2093. doi: 10.1137/110826436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Lu BZ, McCammon JA. J. Chem. Theory Comput. 2007;3:1134. doi: 10.1021/ct700001x. [DOI] [PubMed] [Google Scholar]
- (65).Lu BZ, Zhang DQ, McCammon JA. Journal of Chemical Physics. 2005;122:7. [Google Scholar]
- (66).Rashin AA. Journal of Physical Chemistry. 1989;93:4664. [Google Scholar]
- (67).Vorobjev YN, Grant JA, Scheraga HA. J. Am. Chem. Soc. 1992;114:3189. [Google Scholar]
- (68).Yoon BJ, Lenhoff AM. Journal of Physical Chemistry. 1992;96:3130. [Google Scholar]
- (69).Grant JA, Pickup BT, Nicholls A. J. Comput. Chem. 2001;22:608. [Google Scholar]
- (70).Cai Q, Ye X, Wang J, Luo R. Journal of Chemical Theory and Computation. 2011;7:3608. doi: 10.1021/ct200389p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Wang J, Cai Q, Ye X, Luo R. J. Chem. Theory Comput. 2012;8:2741. doi: 10.1021/ct300341d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Klamt A, Schuurmann G. Journal of the Chemical Society-Perkin Transactions. 1993;2:799. [Google Scholar]
- (73).Li Z, Ito K. The Immersed Interface Method: Numerical Solutions of PDEs Involving Interfaces and Irregular Domains, SIAM Society for Industrial and Applied Mathematics, Philadelphia. 2006 [Google Scholar]
- (74).Cai Q, Wang J, Zhao HK, Luo R. Journal of Chemical Physics. 2009:130. doi: 10.1063/1.3099708. [DOI] [PubMed] [Google Scholar]
- (75).Davis ME, McCammon JA. J. Comput. Chem. 1991;12:909. [Google Scholar]
- (76).Case DA, Darden TA, Cheatham TE,III, Simmerling CL, Wang J, Duke RE, Luo R, Crowley M, Walker RC, Zhang W, Merz KM, Roberts B, Hayik A, Roitberg A, Seabra G, Swails J, Goetz AW, Kolossvai I, Wong KF, Paesani F, Vanicek J, Wolf RM, Liu J, Brozell SR, Steinbrecher T, Gohlke H, Cai Q, Ye X, Wang J, Hsieh M-J, Cui G, Roe DR, Mathews DH, Seetin MG, Salomon-Ferrer R, Sagui C, Babin V, Luchko T, Gusarov S, Kovalenko A, Kollman PA. AMBER. Vol. 12. University of California; 2012. X., W. [Google Scholar]