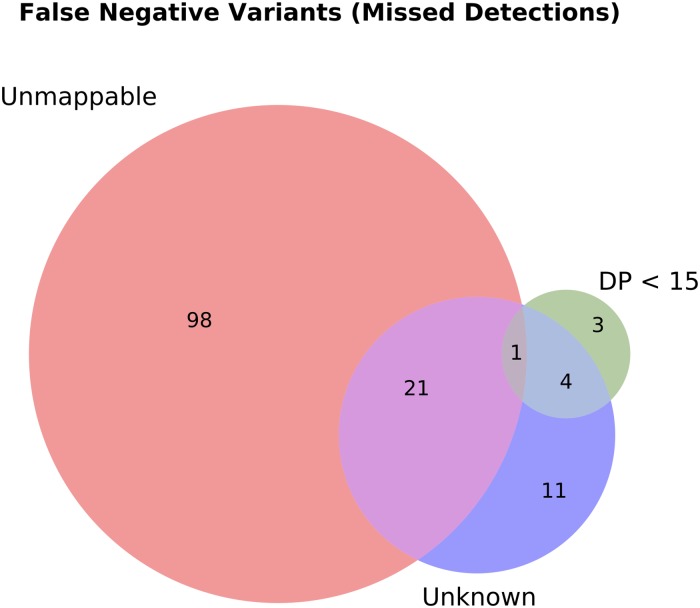
Fig 11. Example false negative variant call diagnosis for a toy dataset: Several hundred variants were introduced into a 10M subset of human chromosome 21. The false negative variants were those that were inserted into the data by NEAT, but were not recovered by a particular variant calling workflow (Novoalign → Haplotype Caller, following GATK best practices).
In this example we see that a majority of the false negatives were due to variants having been inserted into regions that were not uniquely mappable with the simulated read lengths. A lower number of false negatives were due to inadequate coverage (DP).